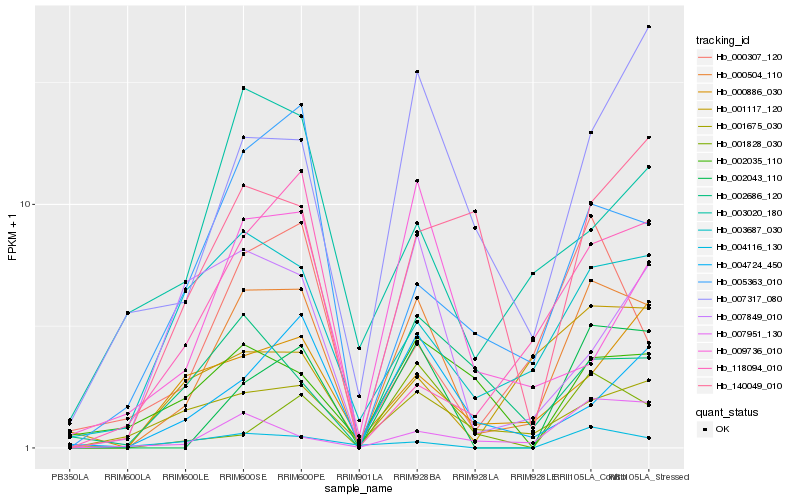
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000504_110 |
0.0 |
- |
- |
F-box/LRR-repeat protein, putative [Ricinus communis] |
| 2 |
Hb_002043_110 |
0.2099386928 |
- |
- |
hypothetical protein POPTR_0001s19500g [Populus trichocarpa] |
| 3 |
Hb_001675_030 |
0.23889643 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_003687_030 |
0.2428241824 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 5 |
Hb_000307_120 |
0.2522665765 |
- |
- |
hypothetical protein B456_002G120900 [Gossypium raimondii] |
| 6 |
Hb_005363_010 |
0.256731506 |
- |
- |
hypothetical protein JCGZ_13835 [Jatropha curcas] |
| 7 |
Hb_004724_450 |
0.2588018204 |
- |
- |
PREDICTED: uncharacterized protein LOC105647069 isoform X1 [Jatropha curcas] |
| 8 |
Hb_004116_130 |
0.2641192894 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 9 |
Hb_007951_130 |
0.2661885987 |
desease resistance |
Gene Name: UBN2_3 |
JHL25H03.6 [Jatropha curcas] |
| 10 |
Hb_001117_120 |
0.2693503378 |
- |
- |
chloride channel clc, putative [Ricinus communis] |
| 11 |
Hb_007317_080 |
0.2716250774 |
- |
- |
PREDICTED: ferredoxin, root R-B2 [Jatropha curcas] |
| 12 |
Hb_000886_030 |
0.2726062045 |
- |
- |
PREDICTED: uncharacterized protein LOC105113410 [Populus euphratica] |
| 13 |
Hb_007849_010 |
0.2767303236 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Populus euphratica] |
| 14 |
Hb_001828_030 |
0.2835017328 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 15 |
Hb_118094_010 |
0.2844602566 |
- |
- |
hypothetical protein POPTR_0011s06595g [Populus trichocarpa] |
| 16 |
Hb_140049_010 |
0.2846877316 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105632009 [Jatropha curcas] |
| 17 |
Hb_002686_120 |
0.2889360785 |
- |
- |
short-chain type dehydrogenase, putative [Ricinus communis] |
| 18 |
Hb_003020_180 |
0.2901960019 |
- |
- |
PREDICTED: probable protein phosphatase 2C 24 [Jatropha curcas] |
| 19 |
Hb_002035_110 |
0.2902527424 |
transcription factor |
TF Family: TRAF |
Regulatory protein NPR1, putative [Ricinus communis] |
| 20 |
Hb_009736_010 |
0.2906054366 |
- |
- |
conserved hypothetical protein [Ricinus communis] |