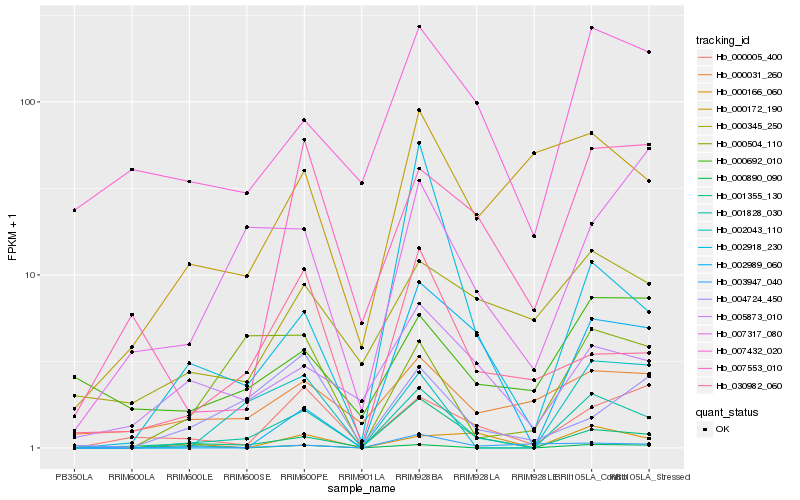
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001828_030 |
0.0 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 2 |
Hb_001355_130 |
0.2448473819 |
- |
- |
PREDICTED: uncharacterized protein LOC105137896 [Populus euphratica] |
| 3 |
Hb_002043_110 |
0.2488884505 |
- |
- |
hypothetical protein POPTR_0001s19500g [Populus trichocarpa] |
| 4 |
Hb_000504_110 |
0.2835017328 |
- |
- |
F-box/LRR-repeat protein, putative [Ricinus communis] |
| 5 |
Hb_000005_400 |
0.2846725063 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 23 isoform X2 [Jatropha curcas] |
| 6 |
Hb_007432_020 |
0.2882896354 |
- |
- |
hypothetical protein POPTR_0010s13310g [Populus trichocarpa] |
| 7 |
Hb_000166_060 |
0.29778503 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_007553_010 |
0.2998505331 |
- |
- |
- |
| 9 |
Hb_005873_010 |
0.3022786029 |
- |
- |
cysteine desulfurylase, putative [Ricinus communis] |
| 10 |
Hb_000890_090 |
0.3079320472 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF086 [Jatropha curcas] |
| 11 |
Hb_030982_060 |
0.3092819429 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002918_230 |
0.3171222516 |
- |
- |
PREDICTED: probable glutathione S-transferase [Gossypium raimondii] |
| 13 |
Hb_000172_190 |
0.3177465362 |
- |
- |
PREDICTED: uncharacterized protein LOC105650824 [Jatropha curcas] |
| 14 |
Hb_003947_040 |
0.3179728252 |
- |
- |
Leucine-rich repeat transmembrane protein kinase protein, putative [Theobroma cacao] |
| 15 |
Hb_007317_080 |
0.3209534211 |
- |
- |
PREDICTED: ferredoxin, root R-B2 [Jatropha curcas] |
| 16 |
Hb_000345_250 |
0.3221169361 |
- |
- |
cullin 1-like protein [Hevea brasiliensis] |
| 17 |
Hb_000031_260 |
0.3242391484 |
- |
- |
hypothetical protein POPTR_0018s03360g [Populus trichocarpa] |
| 18 |
Hb_002989_060 |
0.3257160964 |
- |
- |
hypothetical protein JCGZ_22461 [Jatropha curcas] |
| 19 |
Hb_004724_450 |
0.3266332604 |
- |
- |
PREDICTED: uncharacterized protein LOC105647069 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000692_010 |
0.3270869507 |
- |
- |
PREDICTED: uncharacterized protein LOC105635295 [Jatropha curcas] |