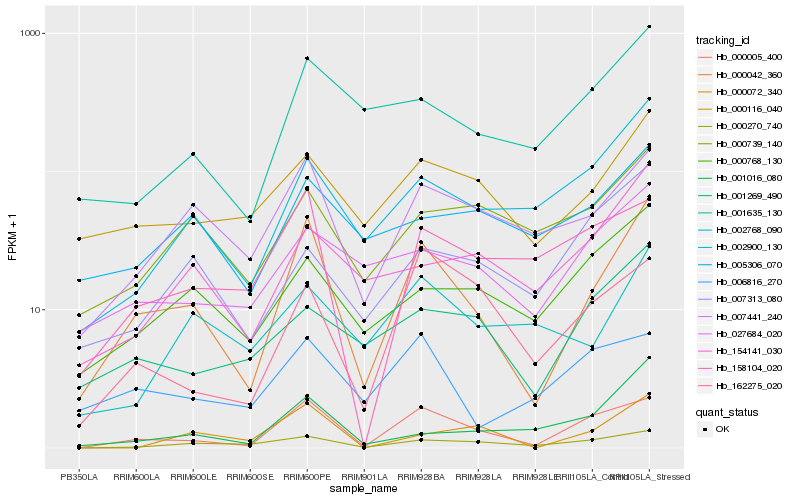
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000042_360 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000005_400 |
0.1626449821 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 23 isoform X2 [Jatropha curcas] |
| 3 |
Hb_007441_240 |
0.2138803828 |
- |
- |
ethphon-induced protein [Hevea brasiliensis] |
| 4 |
Hb_000270_740 |
0.2289674962 |
- |
- |
PREDICTED: probable inactive receptor kinase At2g26730 [Jatropha curcas] |
| 5 |
Hb_000116_040 |
0.2322024135 |
- |
- |
PREDICTED: phosphoserine aminotransferase 1, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_002900_130 |
0.2325028563 |
- |
- |
60S ribosomal L35a-3 -like protein [Gossypium arboreum] |
| 7 |
Hb_001635_130 |
0.2371619733 |
- |
- |
PREDICTED: 60S ribosomal protein L28-2-like [Jatropha curcas] |
| 8 |
Hb_000768_130 |
0.2466664593 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: protein TRI1 [Jatropha curcas] |
| 9 |
Hb_027684_020 |
0.2517094531 |
- |
- |
ribosomal protein S10 (mitochondrion) [Nicotiana tabacum] |
| 10 |
Hb_001269_490 |
0.2534723569 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001016_080 |
0.2539708352 |
- |
- |
hypothetical protein EUGRSUZ_J02452 [Eucalyptus grandis] |
| 12 |
Hb_154141_030 |
0.2541766428 |
- |
- |
PREDICTED: histone H2AX [Populus euphratica] |
| 13 |
Hb_158104_020 |
0.2547815417 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 12 [Jatropha curcas] |
| 14 |
Hb_162275_020 |
0.2589963381 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 15 |
Hb_000072_340 |
0.2605408741 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000739_140 |
0.2614676401 |
- |
- |
PREDICTED: proteasome subunit beta type-1 [Jatropha curcas] |
| 17 |
Hb_006816_270 |
0.2621171907 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002768_090 |
0.2633721934 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 11 [Jatropha curcas] |
| 19 |
Hb_005306_070 |
0.2640109901 |
- |
- |
PREDICTED: coatomer subunit zeta-1-like [Jatropha curcas] |
| 20 |
Hb_007313_080 |
0.2658806249 |
- |
- |
hypothetical protein CICLE_v10002804mg [Citrus clementina] |