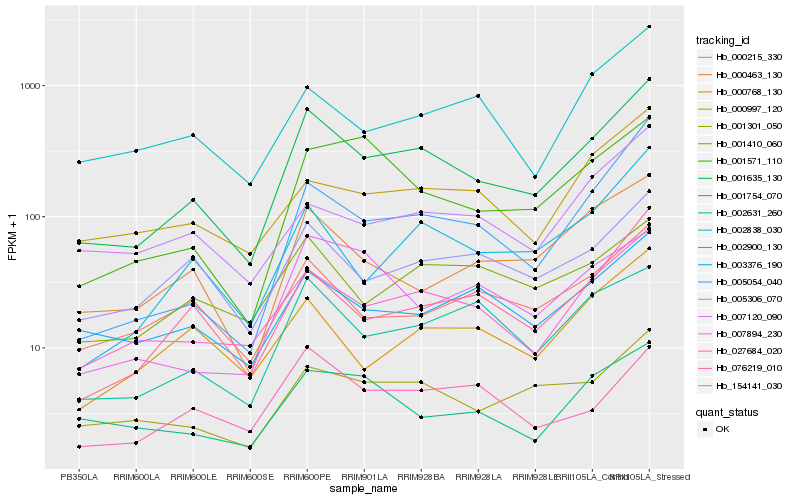
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001635_130 |
0.0 |
- |
- |
PREDICTED: 60S ribosomal protein L28-2-like [Jatropha curcas] |
| 2 |
Hb_027684_020 |
0.1122010814 |
- |
- |
ribosomal protein S10 (mitochondrion) [Nicotiana tabacum] |
| 3 |
Hb_002900_130 |
0.1269157961 |
- |
- |
60S ribosomal L35a-3 -like protein [Gossypium arboreum] |
| 4 |
Hb_154141_030 |
0.1298924058 |
- |
- |
PREDICTED: histone H2AX [Populus euphratica] |
| 5 |
Hb_005054_040 |
0.1314956739 |
- |
- |
PREDICTED: 60S ribosomal protein L35a-1 [Gossypium raimondii] |
| 6 |
Hb_001301_050 |
0.1345337911 |
- |
- |
- |
| 7 |
Hb_002631_260 |
0.1368875705 |
- |
- |
PREDICTED: proteasome subunit alpha type-2-A [Jatropha curcas] |
| 8 |
Hb_000463_130 |
0.1417515238 |
- |
- |
unnamed protein product [Coffea canephora] |
| 9 |
Hb_005306_070 |
0.1422256802 |
- |
- |
PREDICTED: coatomer subunit zeta-1-like [Jatropha curcas] |
| 10 |
Hb_000215_330 |
0.1491452763 |
- |
- |
PREDICTED: AP-4 complex subunit sigma [Jatropha curcas] |
| 11 |
Hb_003376_190 |
0.1510712706 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 12 |
Hb_001410_060 |
0.1515900281 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 8 homolog A-like [Jatropha curcas] |
| 13 |
Hb_000997_120 |
0.15236908 |
- |
- |
40S ribosomal protein S15E [Hevea brasiliensis] |
| 14 |
Hb_000768_130 |
0.1545367007 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: protein TRI1 [Jatropha curcas] |
| 15 |
Hb_001571_110 |
0.1551574926 |
- |
- |
PREDICTED: 60S ribosomal protein L44-like [Gossypium raimondii] |
| 16 |
Hb_002838_030 |
0.1572071212 |
- |
- |
40S ribosomal protein S25A [Hevea brasiliensis] |
| 17 |
Hb_007120_090 |
0.1603765195 |
- |
- |
PREDICTED: 60S ribosomal protein L35 [Jatropha curcas] |
| 18 |
Hb_007894_230 |
0.1615244717 |
- |
- |
30S ribosomal protein S14, putative [Ricinus communis] |
| 19 |
Hb_001754_070 |
0.1615300964 |
- |
- |
hypothetical protein POPTR_0026s00250g [Populus trichocarpa] |
| 20 |
Hb_076219_010 |
0.1626223623 |
- |
- |
PREDICTED: RNA pseudouridine synthase 4, mitochondrial [Jatropha curcas] |