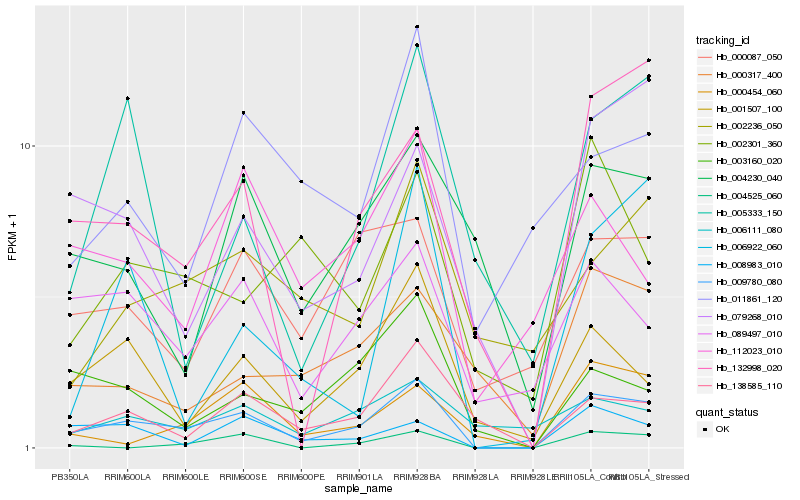
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009780_080 |
0.0 |
- |
- |
PREDICTED: valine--tRNA ligase, mitochondrial isoform X2 [Jatropha curcas] |
| 2 |
Hb_132998_020 |
0.2131164869 |
- |
- |
acyl carrier protein 1, chloroplastic-like [Jatropha curcas] |
| 3 |
Hb_001507_100 |
0.2176033961 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 4 |
Hb_008983_010 |
0.2258624735 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 5 |
Hb_079268_010 |
0.2268740015 |
- |
- |
- |
| 6 |
Hb_089497_010 |
0.2271992491 |
- |
- |
PREDICTED: protein EARLY RESPONSIVE TO DEHYDRATION 15-like isoform X3 [Jatropha curcas] |
| 7 |
Hb_138585_110 |
0.2374783904 |
- |
- |
- |
| 8 |
Hb_000087_050 |
0.23915406 |
- |
- |
hypothetical protein JCGZ_16227 [Jatropha curcas] |
| 9 |
Hb_000454_060 |
0.2444476758 |
- |
- |
Uncharacterized protein TCM_006778 [Theobroma cacao] |
| 10 |
Hb_003160_020 |
0.2456581864 |
- |
- |
PREDICTED: uncharacterized protein LOC105639457 [Jatropha curcas] |
| 11 |
Hb_112023_010 |
0.2511437648 |
- |
- |
hypothetical protein EUGRSUZ_C03826 [Eucalyptus grandis] |
| 12 |
Hb_004525_060 |
0.2512448181 |
- |
- |
- |
| 13 |
Hb_005333_150 |
0.2555608169 |
- |
- |
hypothetical protein POPTR_0002s10520g [Populus trichocarpa] |
| 14 |
Hb_004230_040 |
0.2575015005 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_006922_060 |
0.2586918786 |
- |
- |
calcineurin B, putative [Ricinus communis] |
| 16 |
Hb_006111_080 |
0.2587696915 |
- |
- |
hypothetical protein JCGZ_18070 [Jatropha curcas] |
| 17 |
Hb_002301_360 |
0.259414114 |
- |
- |
hypothetical protein CICLE_v10017252mg [Citrus clementina] |
| 18 |
Hb_002236_050 |
0.2602705067 |
- |
- |
- |
| 19 |
Hb_011861_120 |
0.2614235769 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4 [Jatropha curcas] |
| 20 |
Hb_000317_400 |
0.2616670521 |
transcription factor |
TF Family: zf-HD |
conserved hypothetical protein [Ricinus communis] |