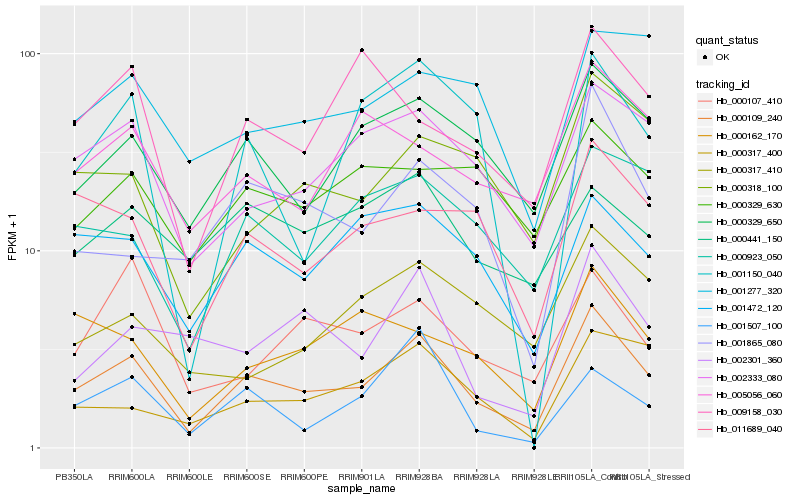
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000109_240 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC102615432 [Citrus sinensis] |
| 2 |
Hb_002333_080 |
0.1496251608 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP20-like [Populus euphratica] |
| 3 |
Hb_000329_650 |
0.1533698401 |
- |
- |
hypothetical protein POPTR_0014s056001g, partial [Populus trichocarpa] |
| 4 |
Hb_000923_050 |
0.1680730821 |
- |
- |
PREDICTED: transcription factor bHLH113 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001472_120 |
0.1683375788 |
- |
- |
- |
| 6 |
Hb_011689_040 |
0.1687021014 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase B5-like [Jatropha curcas] |
| 7 |
Hb_001865_080 |
0.1781101303 |
- |
- |
enoyl-CoA hydratase/isomerase family protein [Populus trichocarpa] |
| 8 |
Hb_000318_100 |
0.1782136967 |
transcription factor |
TF Family: G2-like |
PREDICTED: probable transcription factor KAN3 [Jatropha curcas] |
| 9 |
Hb_000162_170 |
0.1833460629 |
- |
- |
PREDICTED: uncharacterized protein At2g29880-like [Jatropha curcas] |
| 10 |
Hb_002301_360 |
0.1863541525 |
- |
- |
hypothetical protein CICLE_v10017252mg [Citrus clementina] |
| 11 |
Hb_005056_060 |
0.1867222218 |
- |
- |
PREDICTED: prostamide/prostaglandin F synthase isoform X1 [Jatropha curcas] |
| 12 |
Hb_000317_400 |
0.1872605078 |
transcription factor |
TF Family: zf-HD |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000107_410 |
0.1908862116 |
- |
- |
PREDICTED: expansin-A20 [Jatropha curcas] |
| 14 |
Hb_000329_630 |
0.1930414857 |
- |
- |
PREDICTED: nuclear pore complex protein NUP62 [Jatropha curcas] |
| 15 |
Hb_001150_040 |
0.1941269766 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent)-like [Solanum lycopersicum] |
| 16 |
Hb_009158_030 |
0.1947517204 |
transcription factor |
TF Family: HB |
homeobox leucine zipper family protein [Populus trichocarpa] |
| 17 |
Hb_000317_410 |
0.1947565183 |
- |
- |
PREDICTED: UDP-glycosyltransferase 73C5-like [Jatropha curcas] |
| 18 |
Hb_001277_320 |
0.1948536454 |
- |
- |
Stem-specific protein TSJT1, putative [Ricinus communis] |
| 19 |
Hb_001507_100 |
0.1952258805 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 20 |
Hb_000441_150 |
0.1956881942 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, root-type isozyme, chloroplastic isoform X2 [Jatropha curcas] |