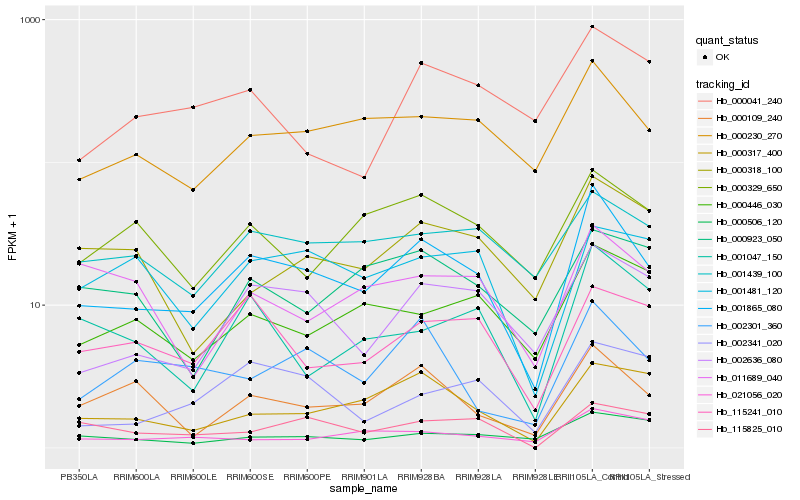
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001865_080 |
0.0 |
- |
- |
enoyl-CoA hydratase/isomerase family protein [Populus trichocarpa] |
| 2 |
Hb_000230_270 |
0.1547312872 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase B5-like [Jatropha curcas] |
| 3 |
Hb_002636_080 |
0.1652089255 |
- |
- |
PREDICTED: sodium-coupled neutral amino acid transporter 3-like [Jatropha curcas] |
| 4 |
Hb_000109_240 |
0.1781101303 |
- |
- |
PREDICTED: uncharacterized protein LOC102615432 [Citrus sinensis] |
| 5 |
Hb_000317_400 |
0.1847327939 |
transcription factor |
TF Family: zf-HD |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002301_360 |
0.1890814617 |
- |
- |
hypothetical protein CICLE_v10017252mg [Citrus clementina] |
| 7 |
Hb_001439_100 |
0.1894707955 |
- |
- |
PREDICTED: SNF1-related protein kinase catalytic subunit alpha KIN10 [Jatropha curcas] |
| 8 |
Hb_001047_150 |
0.1894864736 |
- |
- |
PREDICTED: NF-kappa-B-activating protein [Jatropha curcas] |
| 9 |
Hb_115241_010 |
0.1921415895 |
- |
- |
PREDICTED: protein UPSTREAM OF FLC [Jatropha curcas] |
| 10 |
Hb_000446_030 |
0.1921612134 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 11 |
Hb_000329_650 |
0.1951548977 |
- |
- |
hypothetical protein POPTR_0014s056001g, partial [Populus trichocarpa] |
| 12 |
Hb_021056_020 |
0.2035139035 |
- |
- |
plasma membrane H+-ATPase [Hevea brasiliensis] |
| 13 |
Hb_002341_020 |
0.2037363237 |
- |
- |
PREDICTED: pyruvate decarboxylase 2 [Populus euphratica] |
| 14 |
Hb_115825_010 |
0.2043820756 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 15 |
Hb_000506_120 |
0.2048927241 |
- |
- |
hypothetical protein AALP_AA6G154200 [Arabis alpina] |
| 16 |
Hb_000318_100 |
0.2064032924 |
transcription factor |
TF Family: G2-like |
PREDICTED: probable transcription factor KAN3 [Jatropha curcas] |
| 17 |
Hb_001481_120 |
0.2067244666 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT4-like [Jatropha curcas] |
| 18 |
Hb_000041_240 |
0.2096068465 |
- |
- |
glutaredoxin [Hevea brasiliensis] |
| 19 |
Hb_000923_050 |
0.2110512629 |
- |
- |
PREDICTED: transcription factor bHLH113 isoform X1 [Jatropha curcas] |
| 20 |
Hb_011689_040 |
0.2118281551 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase B5-like [Jatropha curcas] |