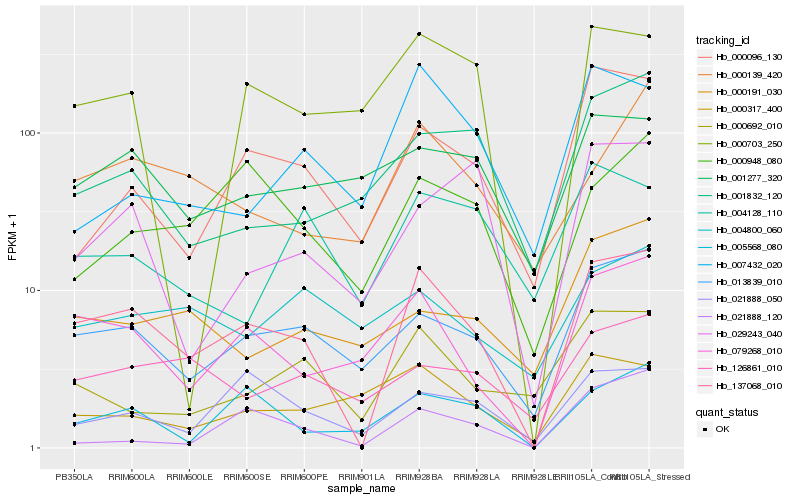
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_137068_010 |
0.0 |
- |
- |
hypothetical protein CISIN_1g003571mg [Citrus sinensis] |
| 2 |
Hb_079268_010 |
0.1846565983 |
- |
- |
- |
| 3 |
Hb_013839_010 |
0.1860007898 |
- |
- |
Angio-associated migratory cell protein, putative [Ricinus communis] |
| 4 |
Hb_005568_080 |
0.2201180602 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000096_130 |
0.2230483363 |
- |
- |
hypothetical protein 23 [Hevea brasiliensis] |
| 6 |
Hb_021888_050 |
0.2250341747 |
- |
- |
PREDICTED: putative CCR4-associated factor 1 homolog 8 [Jatropha curcas] |
| 7 |
Hb_000139_420 |
0.2332805929 |
- |
- |
Stem-specific protein TSJT1, putative [Ricinus communis] |
| 8 |
Hb_000948_080 |
0.2359444375 |
- |
- |
PREDICTED: uncharacterized protein LOC105634692 [Jatropha curcas] |
| 9 |
Hb_000692_010 |
0.2361249353 |
- |
- |
PREDICTED: uncharacterized protein LOC105635295 [Jatropha curcas] |
| 10 |
Hb_126861_010 |
0.238585983 |
- |
- |
hypothetical protein M569_05914, partial [Genlisea aurea] |
| 11 |
Hb_000703_250 |
0.2413391289 |
- |
- |
PREDICTED: protein LIGHT-DEPENDENT SHORT HYPOCOTYLS 10-like [Jatropha curcas] |
| 12 |
Hb_001832_120 |
0.2415938632 |
- |
- |
hypothetical protein PHAVU_003G136800g [Phaseolus vulgaris] |
| 13 |
Hb_001277_320 |
0.2436409932 |
- |
- |
Stem-specific protein TSJT1, putative [Ricinus communis] |
| 14 |
Hb_000317_400 |
0.2508346357 |
transcription factor |
TF Family: zf-HD |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_007432_020 |
0.250913714 |
- |
- |
hypothetical protein POPTR_0010s13310g [Populus trichocarpa] |
| 16 |
Hb_000191_030 |
0.2530596588 |
- |
- |
hypothetical protein POPTR_0012s07830g [Populus trichocarpa] |
| 17 |
Hb_029243_040 |
0.2531526919 |
- |
- |
PREDICTED: alanine--glyoxylate aminotransferase 2 homolog 2, mitochondrial-like [Populus euphratica] |
| 18 |
Hb_021888_120 |
0.2551064506 |
- |
- |
PREDICTED: putative CCR4-associated factor 1 homolog 8 [Jatropha curcas] |
| 19 |
Hb_004128_110 |
0.2574812427 |
- |
- |
glutathione S-transferase L3-like [Jatropha curcas] |
| 20 |
Hb_004800_060 |
0.2582011073 |
- |
- |
PREDICTED: uncharacterized protein LOC105643856 [Jatropha curcas] |