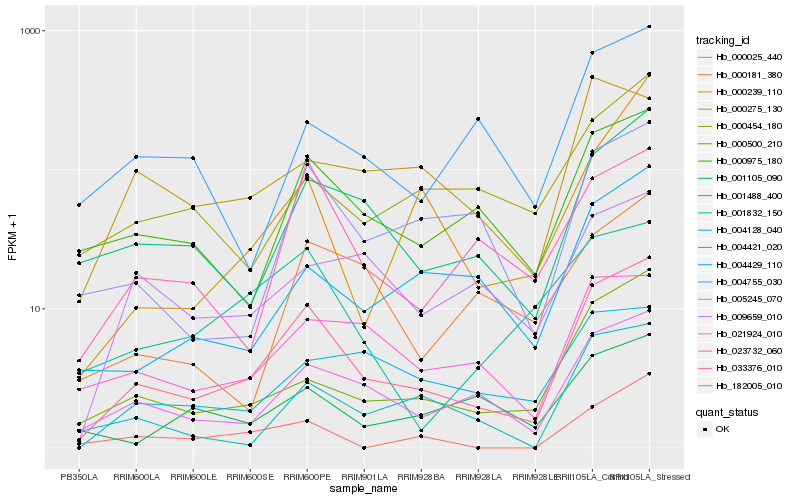
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_182005_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105639166 [Jatropha curcas] |
| 2 |
Hb_021924_010 |
0.1549787717 |
- |
- |
hypothetical protein CISIN_1g0405342mg, partial [Citrus sinensis] |
| 3 |
Hb_001488_400 |
0.1762749011 |
- |
- |
hypothetical protein POPTR_0018s12030g [Populus trichocarpa] |
| 4 |
Hb_000500_210 |
0.177735894 |
- |
- |
- |
| 5 |
Hb_000975_180 |
0.1829464243 |
- |
- |
Uncharacterized protein TCM_041879 [Theobroma cacao] |
| 6 |
Hb_005245_070 |
0.1961109842 |
- |
- |
PREDICTED: uncharacterized protein LOC105643658 [Jatropha curcas] |
| 7 |
Hb_033376_010 |
0.1967113437 |
- |
- |
hypothetical protein POPTR_0008s02960g [Populus trichocarpa] |
| 8 |
Hb_004128_040 |
0.2017774962 |
- |
- |
- |
| 9 |
Hb_004429_110 |
0.2052235306 |
- |
- |
PREDICTED: protein SPIRAL1-like 1 [Populus euphratica] |
| 10 |
Hb_023732_060 |
0.2055806662 |
- |
- |
PREDICTED: uncharacterized protein LOC105637567 [Jatropha curcas] |
| 11 |
Hb_001832_150 |
0.2145573318 |
- |
- |
PREDICTED: uncharacterized protein LOC105637882 [Jatropha curcas] |
| 12 |
Hb_004421_020 |
0.2201108673 |
- |
- |
- |
| 13 |
Hb_000025_440 |
0.2206684515 |
- |
- |
PREDICTED: ankyrin repeat-containing protein P16F5.05c-like [Populus euphratica] |
| 14 |
Hb_000454_180 |
0.2218445021 |
- |
- |
PREDICTED: uncharacterized protein LOC105134496 [Populus euphratica] |
| 15 |
Hb_004755_030 |
0.2232027333 |
- |
- |
EG964 [Manihot glaziovii] |
| 16 |
Hb_000181_380 |
0.2243258193 |
- |
- |
PREDICTED: cytochrome c oxidase copper chaperone 1 [Jatropha curcas] |
| 17 |
Hb_009659_010 |
0.2251976416 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 9 [Vitis vinifera] |
| 18 |
Hb_000239_110 |
0.2259085379 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 1-like [Jatropha curcas] |
| 19 |
Hb_001105_090 |
0.2275029297 |
transcription factor |
TF Family: C3H |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000275_130 |
0.2281865939 |
- |
- |
hypothetical protein CICLE_v10003066mg [Citrus clementina] |