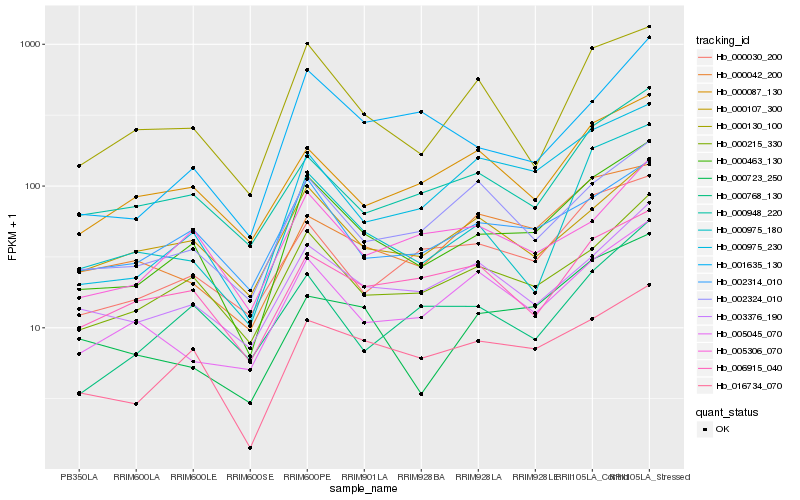
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000463_130 |
0.0 |
- |
- |
unnamed protein product [Coffea canephora] |
| 2 |
Hb_000215_330 |
0.1047341315 |
- |
- |
PREDICTED: AP-4 complex subunit sigma [Jatropha curcas] |
| 3 |
Hb_016734_070 |
0.1132857706 |
- |
- |
hypothetical_protein [Oryza brachyantha] |
| 4 |
Hb_002314_010 |
0.1237136729 |
- |
- |
hypothetical protein JCGZ_15712 [Jatropha curcas] |
| 5 |
Hb_000130_100 |
0.1296193715 |
- |
- |
hypothetical protein CICLE_v10022662mg [Citrus clementina] |
| 6 |
Hb_000975_230 |
0.1307636604 |
- |
- |
hypothetical protein ZEAMMB73_864543 [Zea mays] |
| 7 |
Hb_003376_190 |
0.131012901 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 8 |
Hb_000975_180 |
0.1313342781 |
- |
- |
Uncharacterized protein TCM_041879 [Theobroma cacao] |
| 9 |
Hb_000030_200 |
0.1328131036 |
- |
- |
PREDICTED: early nodulin-93-like [Populus euphratica] |
| 10 |
Hb_000948_220 |
0.1332409784 |
- |
- |
hypothetical protein CICLE_v10013189mg [Citrus clementina] |
| 11 |
Hb_002324_010 |
0.135854299 |
- |
- |
PREDICTED: proteasome subunit alpha type-7 [Jatropha curcas] |
| 12 |
Hb_000107_300 |
0.1359502993 |
- |
- |
PREDICTED: AP-1 complex subunit sigma-2 [Jatropha curcas] |
| 13 |
Hb_005045_070 |
0.138082258 |
- |
- |
PREDICTED: deSI-like protein At4g17486 [Jatropha curcas] |
| 14 |
Hb_000723_250 |
0.1387046924 |
- |
- |
hypothetical protein JCGZ_16520 [Jatropha curcas] |
| 15 |
Hb_005306_070 |
0.1399770579 |
- |
- |
PREDICTED: coatomer subunit zeta-1-like [Jatropha curcas] |
| 16 |
Hb_001635_130 |
0.1417515238 |
- |
- |
PREDICTED: 60S ribosomal protein L28-2-like [Jatropha curcas] |
| 17 |
Hb_000087_130 |
0.1419750463 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 6, mitochondrial [Jatropha curcas] |
| 18 |
Hb_000042_200 |
0.1425295854 |
- |
- |
PREDICTED: uncharacterized protein LOC100252136 isoform X1 [Vitis vinifera] |
| 19 |
Hb_006915_040 |
0.1460459387 |
- |
- |
PREDICTED: prefoldin subunit 6 [Eucalyptus grandis] |
| 20 |
Hb_000768_130 |
0.1464241922 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: protein TRI1 [Jatropha curcas] |