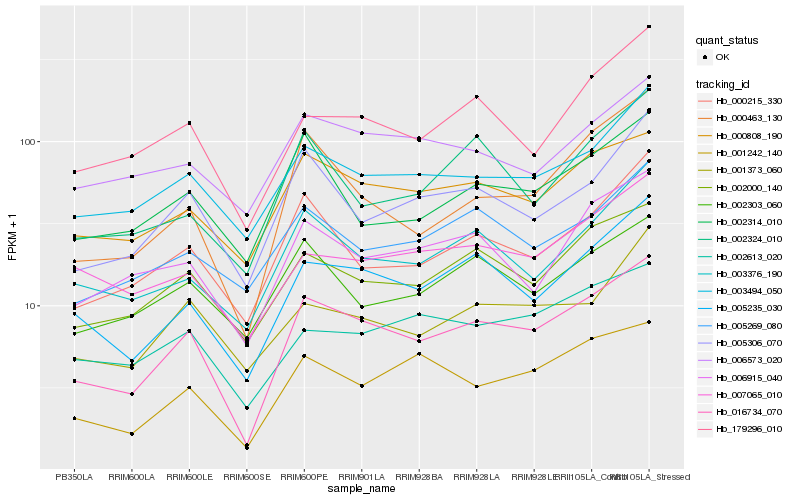
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_016734_070 |
0.0 |
- |
- |
hypothetical_protein [Oryza brachyantha] |
| 2 |
Hb_005235_030 |
0.0925714815 |
- |
- |
PREDICTED: dolichol phosphate-mannose biosynthesis regulatory protein [Eucalyptus grandis] |
| 3 |
Hb_002000_140 |
0.1081614842 |
- |
- |
PREDICTED: thioredoxin Y2, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000808_190 |
0.1110388592 |
- |
- |
PREDICTED: GTP-binding protein SAR1A-like [Eucalyptus grandis] |
| 5 |
Hb_000463_130 |
0.1132857706 |
- |
- |
unnamed protein product [Coffea canephora] |
| 6 |
Hb_001242_140 |
0.1146477212 |
- |
- |
mitochondrial carrier protein, putative [Ricinus communis] |
| 7 |
Hb_000215_330 |
0.114739905 |
- |
- |
PREDICTED: AP-4 complex subunit sigma [Jatropha curcas] |
| 8 |
Hb_005306_070 |
0.115028047 |
- |
- |
PREDICTED: coatomer subunit zeta-1-like [Jatropha curcas] |
| 9 |
Hb_003494_050 |
0.1159047729 |
- |
- |
hypothetical protein PRUPE_ppa013459mg [Prunus persica] |
| 10 |
Hb_003376_190 |
0.117270764 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 11 |
Hb_006915_040 |
0.118350822 |
- |
- |
PREDICTED: prefoldin subunit 6 [Eucalyptus grandis] |
| 12 |
Hb_179296_010 |
0.1205086532 |
- |
- |
hypothetical protein B456_006G094200 [Gossypium raimondii] |
| 13 |
Hb_002613_020 |
0.1274984168 |
- |
- |
ADP-ribose pyrophosphatase, putative [Ricinus communis] |
| 14 |
Hb_002314_010 |
0.1282127562 |
- |
- |
hypothetical protein JCGZ_15712 [Jatropha curcas] |
| 15 |
Hb_005269_080 |
0.1285709428 |
- |
- |
protein with unknown function [Ricinus communis] |
| 16 |
Hb_001373_060 |
0.1288838863 |
- |
- |
PREDICTED: structure-specific endonuclease subunit slx1 [Jatropha curcas] |
| 17 |
Hb_007065_010 |
0.1314797539 |
- |
- |
hypothetical protein Csa_3G379730 [Cucumis sativus] |
| 18 |
Hb_002324_010 |
0.1319697321 |
- |
- |
PREDICTED: proteasome subunit alpha type-7 [Jatropha curcas] |
| 19 |
Hb_002303_060 |
0.1323122506 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_006573_020 |
0.1330924003 |
- |
- |
PREDICTED: 40S ribosomal protein S10-like [Jatropha curcas] |