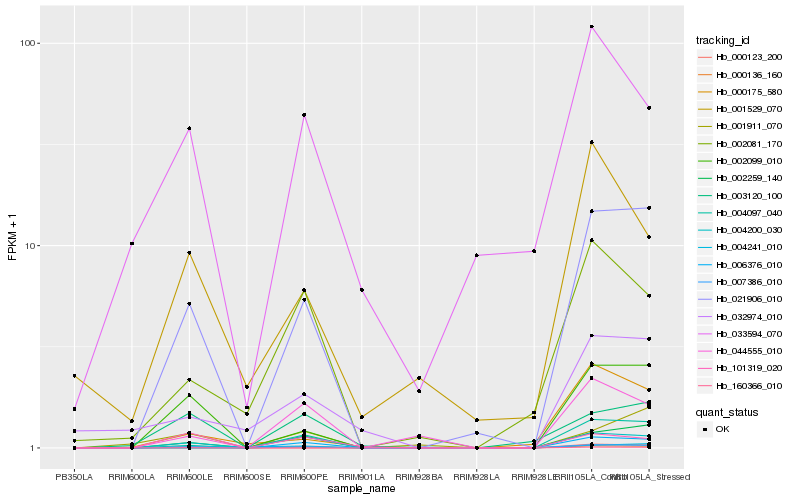
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007386_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 2 |
Hb_021906_010 |
0.0860295726 |
- |
- |
PREDICTED: cysteine proteinase inhibitor-like [Populus euphratica] |
| 3 |
Hb_004200_030 |
0.1210046017 |
- |
- |
PREDICTED: SWR1 complex subunit 6-like isoform X1 [Citrus sinensis] |
| 4 |
Hb_002099_010 |
0.1491936488 |
- |
- |
cysteine proteinase inhibitor 5-like [Cucumis sativus] |
| 5 |
Hb_002259_140 |
0.150835538 |
- |
- |
- |
| 6 |
Hb_004097_040 |
0.1840140026 |
- |
- |
PREDICTED: aspartate--tRNA ligase, mitochondrial [Jatropha curcas] |
| 7 |
Hb_044555_010 |
0.235250607 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X3 [Jatropha curcas] |
| 8 |
Hb_002081_170 |
0.2427684764 |
- |
- |
Flavonol 3-sulfotransferase, putative [Ricinus communis] |
| 9 |
Hb_101319_020 |
0.2428714387 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 10 |
Hb_003120_100 |
0.2438518036 |
- |
- |
PREDICTED: GDSL esterase/lipase EXL3-like [Jatropha curcas] |
| 11 |
Hb_000175_580 |
0.2546932373 |
transcription factor, desease resistance |
TF Family: C3H, Gene Name: zf-CCCH |
hypothetical protein POPTR_0013s00890g [Populus trichocarpa] |
| 12 |
Hb_004241_010 |
0.2607899828 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 13 |
Hb_001529_070 |
0.26599334 |
transcription factor |
TF Family: AP2 |
hypothetical protein POPTR_0018s09900g [Populus trichocarpa] |
| 14 |
Hb_006376_010 |
0.2745507038 |
- |
- |
PREDICTED: uncharacterized protein LOC104090718 [Nicotiana tomentosiformis] |
| 15 |
Hb_032974_010 |
0.2746744719 |
- |
- |
- |
| 16 |
Hb_033594_070 |
0.2747128151 |
- |
- |
PREDICTED: uncharacterized protein LOC105787685 isoform X1 [Gossypium raimondii] |
| 17 |
Hb_000123_200 |
0.2754185272 |
- |
- |
PREDICTED: WAT1-related protein At2g39510 [Jatropha curcas] |
| 18 |
Hb_160366_010 |
0.2755442383 |
- |
- |
PREDICTED: uncharacterized protein LOC104894399 [Beta vulgaris subsp. vulgaris] |
| 19 |
Hb_001911_070 |
0.2787955784 |
- |
- |
- |
| 20 |
Hb_000136_160 |
0.2861629967 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |