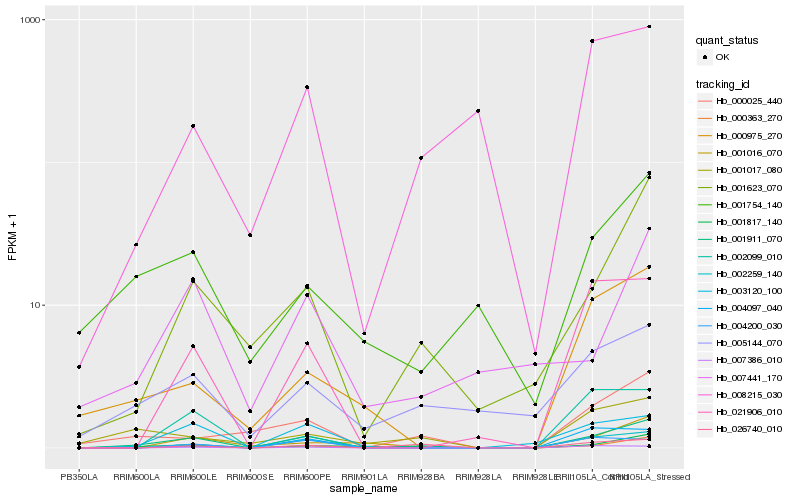
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001911_070 |
0.0 |
- |
- |
- |
| 2 |
Hb_001817_140 |
0.2170414164 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 3 |
Hb_002259_140 |
0.2170868969 |
- |
- |
- |
| 4 |
Hb_001623_070 |
0.2438818974 |
- |
- |
cytochrome B5 isoform 1, putative [Ricinus communis] |
| 5 |
Hb_003120_100 |
0.2495400967 |
- |
- |
PREDICTED: GDSL esterase/lipase EXL3-like [Jatropha curcas] |
| 6 |
Hb_021906_010 |
0.2625022853 |
- |
- |
PREDICTED: cysteine proteinase inhibitor-like [Populus euphratica] |
| 7 |
Hb_000025_440 |
0.2628189921 |
- |
- |
PREDICTED: ankyrin repeat-containing protein P16F5.05c-like [Populus euphratica] |
| 8 |
Hb_026740_010 |
0.2673417227 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 [Jatropha curcas] |
| 9 |
Hb_000363_270 |
0.2722897021 |
- |
- |
hypothetical protein PRUPE_ppa024846mg [Prunus persica] |
| 10 |
Hb_007386_010 |
0.2787955784 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 11 |
Hb_005144_070 |
0.281526423 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_004200_030 |
0.2849962277 |
- |
- |
PREDICTED: SWR1 complex subunit 6-like isoform X1 [Citrus sinensis] |
| 13 |
Hb_000975_270 |
0.2890236618 |
- |
- |
PREDICTED: histone H2B.11-like [Phoenix dactylifera] |
| 14 |
Hb_001016_070 |
0.2901710234 |
- |
- |
PREDICTED: putative MO25-like protein At5g47540 [Jatropha curcas] |
| 15 |
Hb_001754_140 |
0.2916520601 |
- |
- |
PREDICTED: uncharacterized protein LOC105643419 [Jatropha curcas] |
| 16 |
Hb_001017_080 |
0.2922678176 |
- |
- |
hypothetical protein POPTR_0010s06020g [Populus trichocarpa] |
| 17 |
Hb_008215_030 |
0.2925827382 |
- |
- |
hypothetical protein 3 [Hevea brasiliensis] |
| 18 |
Hb_007441_170 |
0.2973039741 |
- |
- |
PREDICTED: rac-like GTP-binding protein RAC2 [Jatropha curcas] |
| 19 |
Hb_002099_010 |
0.2992749733 |
- |
- |
cysteine proteinase inhibitor 5-like [Cucumis sativus] |
| 20 |
Hb_004097_040 |
0.2999115383 |
- |
- |
PREDICTED: aspartate--tRNA ligase, mitochondrial [Jatropha curcas] |