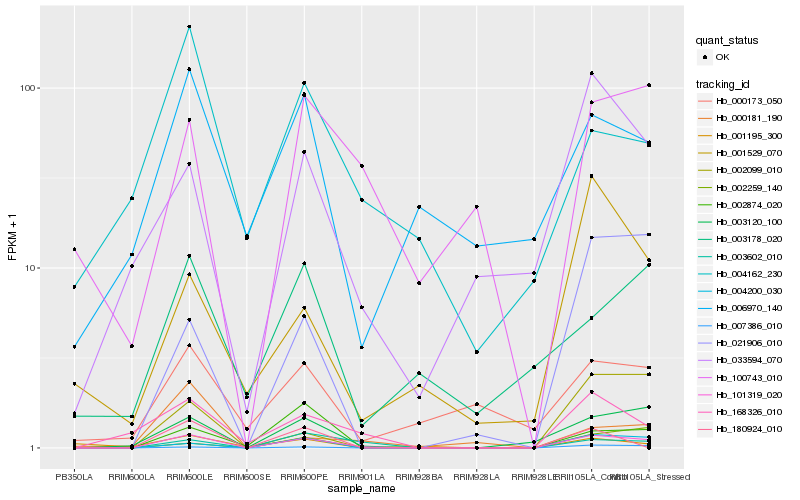
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_101319_020 |
0.0 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 2 |
Hb_003120_100 |
0.2127892277 |
- |
- |
PREDICTED: GDSL esterase/lipase EXL3-like [Jatropha curcas] |
| 3 |
Hb_004200_030 |
0.2301428198 |
- |
- |
PREDICTED: SWR1 complex subunit 6-like isoform X1 [Citrus sinensis] |
| 4 |
Hb_007386_010 |
0.2428714387 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 5 |
Hb_003602_010 |
0.2490548665 |
- |
- |
serine hydroxymethyltransferase, partial [Populus alba] |
| 6 |
Hb_168326_010 |
0.2499983919 |
- |
- |
PREDICTED: alpha-L-fucosidase 2-like isoform X2 [Populus euphratica] |
| 7 |
Hb_002874_020 |
0.2841320814 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105637459 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002099_010 |
0.2947036161 |
- |
- |
cysteine proteinase inhibitor 5-like [Cucumis sativus] |
| 9 |
Hb_033594_070 |
0.2965014827 |
- |
- |
PREDICTED: uncharacterized protein LOC105787685 isoform X1 [Gossypium raimondii] |
| 10 |
Hb_021906_010 |
0.2967189655 |
- |
- |
PREDICTED: cysteine proteinase inhibitor-like [Populus euphratica] |
| 11 |
Hb_100743_010 |
0.3029242214 |
- |
- |
hypothetical protein JCGZ_17104 [Jatropha curcas] |
| 12 |
Hb_002259_140 |
0.3039509011 |
- |
- |
- |
| 13 |
Hb_000173_050 |
0.3173179774 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 14 |
Hb_006970_140 |
0.3289555688 |
- |
- |
PREDICTED: histone H3.2-like [Solanum lycopersicum] |
| 15 |
Hb_003178_020 |
0.3316043932 |
transcription factor |
TF Family: B3 |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001529_070 |
0.333955745 |
transcription factor |
TF Family: AP2 |
hypothetical protein POPTR_0018s09900g [Populus trichocarpa] |
| 17 |
Hb_180924_010 |
0.3360740181 |
- |
- |
CPI-4 [Morus alba var. atropurpurea] |
| 18 |
Hb_004162_230 |
0.3371839395 |
- |
- |
PREDICTED: histone H2B [Jatropha curcas] |
| 19 |
Hb_001195_300 |
0.338212738 |
- |
- |
PREDICTED: DUF21 domain-containing protein At1g47330 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000181_190 |
0.3405104695 |
- |
- |
PREDICTED: probable protein phosphatase 2C 39 [Jatropha curcas] |