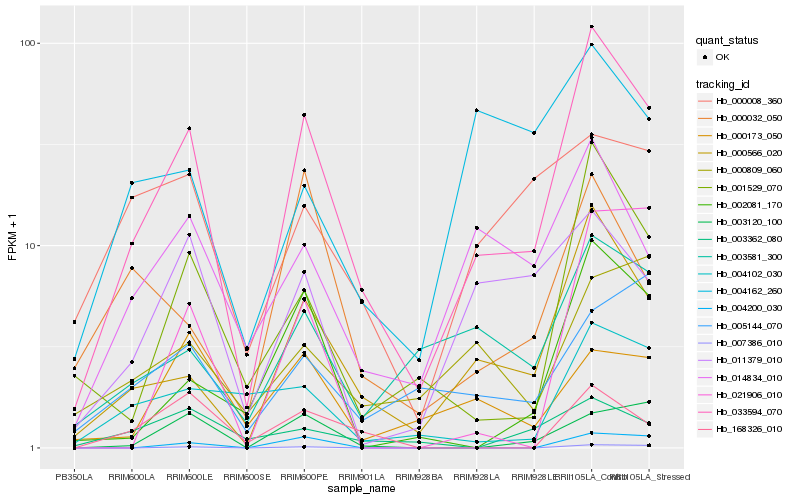
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033594_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105787685 isoform X1 [Gossypium raimondii] |
| 2 |
Hb_000566_020 |
0.1599256636 |
- |
- |
hypothetical protein POPTR_0001s13520g [Populus trichocarpa] |
| 3 |
Hb_001529_070 |
0.2194976451 |
transcription factor |
TF Family: AP2 |
hypothetical protein POPTR_0018s09900g [Populus trichocarpa] |
| 4 |
Hb_014834_010 |
0.2393632657 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT4 [Jatropha curcas] |
| 5 |
Hb_002081_170 |
0.2418212488 |
- |
- |
Flavonol 3-sulfotransferase, putative [Ricinus communis] |
| 6 |
Hb_003581_300 |
0.2459725826 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105630404 isoform X1 [Jatropha curcas] |
| 7 |
Hb_004102_030 |
0.2608039882 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g63020 [Jatropha curcas] |
| 8 |
Hb_011379_010 |
0.2669962436 |
- |
- |
Thylakoid lumenal 25.6 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 9 |
Hb_003120_100 |
0.270532089 |
- |
- |
PREDICTED: GDSL esterase/lipase EXL3-like [Jatropha curcas] |
| 10 |
Hb_007386_010 |
0.2747128151 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 11 |
Hb_000032_050 |
0.2821105078 |
- |
- |
hypothetical protein RCOM_1691130 [Ricinus communis] |
| 12 |
Hb_004200_030 |
0.2834314794 |
- |
- |
PREDICTED: SWR1 complex subunit 6-like isoform X1 [Citrus sinensis] |
| 13 |
Hb_000809_060 |
0.2844026955 |
transcription factor |
TF Family: LOB |
LOB domain-containing 36 -like protein [Gossypium arboreum] |
| 14 |
Hb_168326_010 |
0.2844858016 |
- |
- |
PREDICTED: alpha-L-fucosidase 2-like isoform X2 [Populus euphratica] |
| 15 |
Hb_000173_050 |
0.286633035 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 16 |
Hb_005144_070 |
0.287290371 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000008_360 |
0.2873418792 |
- |
- |
hypothetical protein POPTR_0004s22050g [Populus trichocarpa] |
| 18 |
Hb_003362_080 |
0.2889488313 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 19 |
Hb_021906_010 |
0.2945640017 |
- |
- |
PREDICTED: cysteine proteinase inhibitor-like [Populus euphratica] |
| 20 |
Hb_004162_260 |
0.2945681181 |
transcription factor |
TF Family: C2H2 |
conserved hypothetical protein [Ricinus communis] |