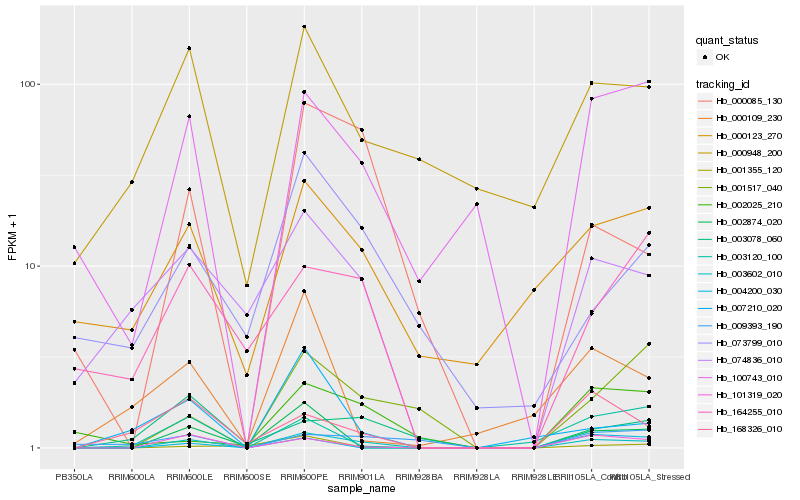
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003602_010 |
0.0 |
- |
- |
serine hydroxymethyltransferase, partial [Populus alba] |
| 2 |
Hb_101319_020 |
0.2490548665 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 3 |
Hb_000085_130 |
0.2741419202 |
- |
- |
- |
| 4 |
Hb_100743_010 |
0.2758840959 |
- |
- |
hypothetical protein JCGZ_17104 [Jatropha curcas] |
| 5 |
Hb_002874_020 |
0.2838620464 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105637459 isoform X1 [Jatropha curcas] |
| 6 |
Hb_074836_010 |
0.2891993007 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_168326_010 |
0.3140959946 |
- |
- |
PREDICTED: alpha-L-fucosidase 2-like isoform X2 [Populus euphratica] |
| 8 |
Hb_000123_270 |
0.3154538446 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000948_200 |
0.3181953067 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 10 |
Hb_001517_040 |
0.318315311 |
- |
- |
- |
| 11 |
Hb_003120_100 |
0.3196520146 |
- |
- |
PREDICTED: GDSL esterase/lipase EXL3-like [Jatropha curcas] |
| 12 |
Hb_007210_020 |
0.3239242961 |
- |
- |
PREDICTED: hexaprenyldihydroxybenzoate methyltransferase, mitochondrial-like [Solanum tuberosum] |
| 13 |
Hb_164255_010 |
0.3248682073 |
- |
- |
PREDICTED: aspartate-semialdehyde dehydrogenase [Jatropha curcas] |
| 14 |
Hb_001355_120 |
0.3274978529 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 15 |
Hb_004200_030 |
0.3296146664 |
- |
- |
PREDICTED: SWR1 complex subunit 6-like isoform X1 [Citrus sinensis] |
| 16 |
Hb_073799_010 |
0.3321434036 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000109_230 |
0.3359269785 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_003078_060 |
0.338153839 |
- |
- |
60S ribosomal protein L32, putative [Ricinus communis] |
| 19 |
Hb_002025_210 |
0.3405284064 |
- |
- |
- |
| 20 |
Hb_009393_190 |
0.3415690747 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase AIP2-like [Jatropha curcas] |