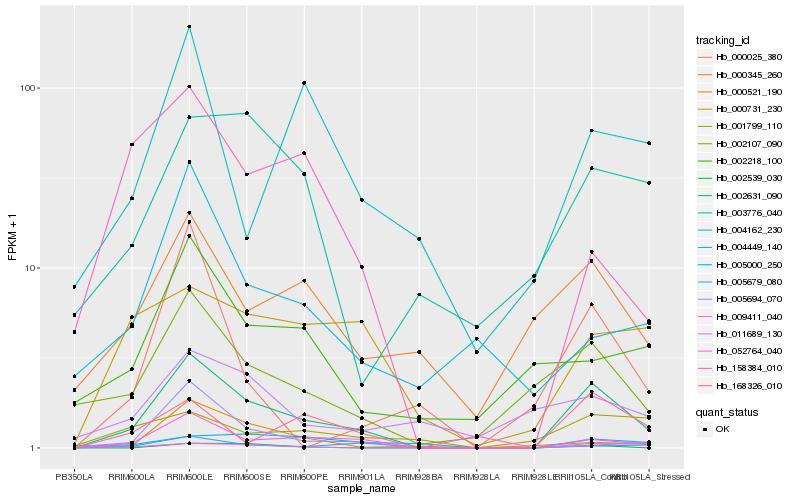
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002631_090 |
0.0 |
- |
- |
- |
| 2 |
Hb_002107_090 |
0.2669917686 |
transcription factor |
TF Family: bZIP |
Basic-leucine zipper transcription factor family protein, putative [Theobroma cacao] |
| 3 |
Hb_168326_010 |
0.2819462392 |
- |
- |
PREDICTED: alpha-L-fucosidase 2-like isoform X2 [Populus euphratica] |
| 4 |
Hb_005679_080 |
0.2984104125 |
- |
- |
PREDICTED: PH-interacting protein-like [Populus euphratica] |
| 5 |
Hb_000345_260 |
0.3001115037 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Citrus sinensis] |
| 6 |
Hb_002539_030 |
0.3146516093 |
- |
- |
PREDICTED: uncharacterized protein LOC105630961 [Jatropha curcas] |
| 7 |
Hb_000731_230 |
0.3156055421 |
- |
- |
PREDICTED: abnormal spindle-like microcephaly-associated protein homolog [Jatropha curcas] |
| 8 |
Hb_052764_040 |
0.3185367535 |
- |
- |
syntaxin 125 family protein [Populus trichocarpa] |
| 9 |
Hb_000521_190 |
0.3207408292 |
desease resistance |
Gene Name: TIR |
hypothetical protein JCGZ_25500 [Jatropha curcas] |
| 10 |
Hb_011689_130 |
0.3218393577 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK2 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002218_100 |
0.3225723774 |
- |
- |
PREDICTED: uncharacterized protein At2g34160 [Jatropha curcas] |
| 12 |
Hb_003776_040 |
0.3237806047 |
- |
- |
PREDICTED: RING finger protein 165-like isoform X2 [Populus euphratica] |
| 13 |
Hb_004162_230 |
0.3254885112 |
- |
- |
PREDICTED: histone H2B [Jatropha curcas] |
| 14 |
Hb_158384_010 |
0.327174316 |
- |
- |
poly(A) polymerase, putative [Ricinus communis] |
| 15 |
Hb_000025_380 |
0.3276051231 |
- |
- |
hypothetical protein JCGZ_22787 [Jatropha curcas] |
| 16 |
Hb_004449_140 |
0.3285857904 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor GAMYB-like [Jatropha curcas] |
| 17 |
Hb_005694_070 |
0.3290792393 |
- |
- |
kinase, putative [Ricinus communis] |
| 18 |
Hb_009411_040 |
0.3301549418 |
- |
- |
- |
| 19 |
Hb_001799_110 |
0.3349942806 |
- |
- |
PREDICTED: glycine--tRNA ligase 2, chloroplastic/mitochondrial isoform X2 [Jatropha curcas] |
| 20 |
Hb_005000_250 |
0.336803703 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 4, mitochondrial [Vitis vinifera] |