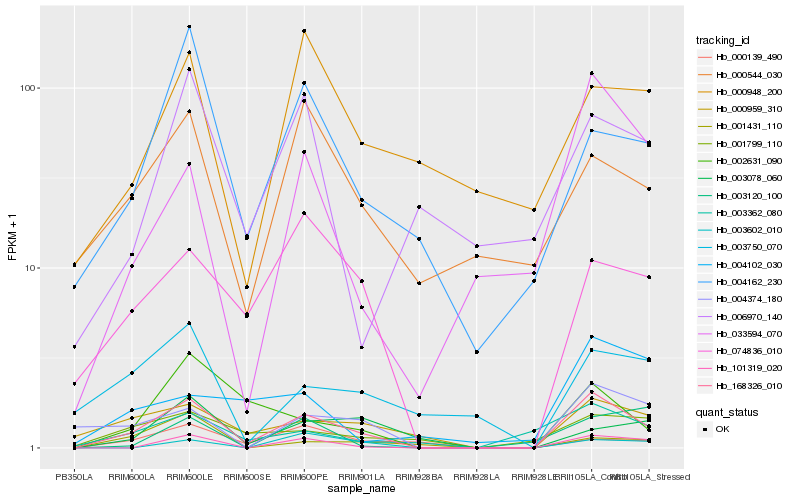
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_168326_010 |
0.0 |
- |
- |
PREDICTED: alpha-L-fucosidase 2-like isoform X2 [Populus euphratica] |
| 2 |
Hb_101319_020 |
0.2499983919 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 3 |
Hb_074836_010 |
0.2618339949 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_004374_180 |
0.2660723386 |
- |
- |
GMP synthase, putative [Ricinus communis] |
| 5 |
Hb_000959_310 |
0.2686220675 |
- |
- |
PREDICTED: WD repeat domain-containing protein 83 [Jatropha curcas] |
| 6 |
Hb_002631_090 |
0.2819462392 |
- |
- |
- |
| 7 |
Hb_033594_070 |
0.2844858016 |
- |
- |
PREDICTED: uncharacterized protein LOC105787685 isoform X1 [Gossypium raimondii] |
| 8 |
Hb_003362_080 |
0.2873722297 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 9 |
Hb_004162_230 |
0.2916449823 |
- |
- |
PREDICTED: histone H2B [Jatropha curcas] |
| 10 |
Hb_001799_110 |
0.3005994711 |
- |
- |
PREDICTED: glycine--tRNA ligase 2, chloroplastic/mitochondrial isoform X2 [Jatropha curcas] |
| 11 |
Hb_000139_490 |
0.3049272653 |
- |
- |
PREDICTED: transcription factor MUTE [Jatropha curcas] |
| 12 |
Hb_003750_070 |
0.3074239081 |
- |
- |
PREDICTED: psbQ-like protein 3, chloroplastic [Jatropha curcas] |
| 13 |
Hb_003078_060 |
0.3137814141 |
- |
- |
60S ribosomal protein L32, putative [Ricinus communis] |
| 14 |
Hb_003602_010 |
0.3140959946 |
- |
- |
serine hydroxymethyltransferase, partial [Populus alba] |
| 15 |
Hb_000544_030 |
0.3148909248 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_006970_140 |
0.3303181432 |
- |
- |
PREDICTED: histone H3.2-like [Solanum lycopersicum] |
| 17 |
Hb_001431_110 |
0.330333886 |
- |
- |
glycerol-3-phosphate transporter family protein [Populus trichocarpa] |
| 18 |
Hb_004102_030 |
0.3312936955 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g63020 [Jatropha curcas] |
| 19 |
Hb_000948_200 |
0.3316601196 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 20 |
Hb_003120_100 |
0.331672576 |
- |
- |
PREDICTED: GDSL esterase/lipase EXL3-like [Jatropha curcas] |