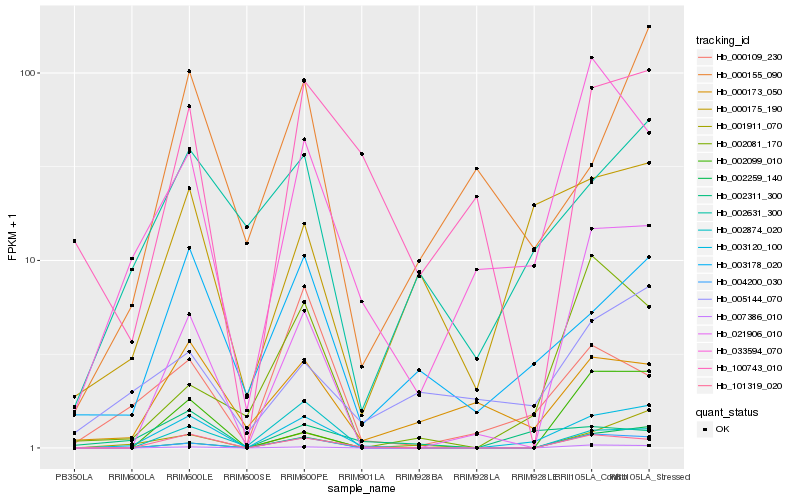
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003120_100 |
0.0 |
- |
- |
PREDICTED: GDSL esterase/lipase EXL3-like [Jatropha curcas] |
| 2 |
Hb_101319_020 |
0.2127892277 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 3 |
Hb_004200_030 |
0.2312990223 |
- |
- |
PREDICTED: SWR1 complex subunit 6-like isoform X1 [Citrus sinensis] |
| 4 |
Hb_002259_140 |
0.2399083232 |
- |
- |
- |
| 5 |
Hb_007386_010 |
0.2438518036 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 6 |
Hb_001911_070 |
0.2495400967 |
- |
- |
- |
| 7 |
Hb_003178_020 |
0.2537067748 |
transcription factor |
TF Family: B3 |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_021906_010 |
0.2659382765 |
- |
- |
PREDICTED: cysteine proteinase inhibitor-like [Populus euphratica] |
| 9 |
Hb_033594_070 |
0.270532089 |
- |
- |
PREDICTED: uncharacterized protein LOC105787685 isoform X1 [Gossypium raimondii] |
| 10 |
Hb_002631_300 |
0.282842725 |
- |
- |
PREDICTED: uncharacterized protein LOC105646181 [Jatropha curcas] |
| 11 |
Hb_000175_190 |
0.285473101 |
- |
- |
PREDICTED: phosphatidylcholine:diacylglycerol cholinephosphotransferase 1-like [Jatropha curcas] |
| 12 |
Hb_002099_010 |
0.2892713031 |
- |
- |
cysteine proteinase inhibitor 5-like [Cucumis sativus] |
| 13 |
Hb_000173_050 |
0.2966800175 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 14 |
Hb_000155_090 |
0.2982600617 |
- |
- |
PREDICTED: glutaredoxin-C9 [Jatropha curcas] |
| 15 |
Hb_005144_070 |
0.3081974795 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002874_020 |
0.3082684417 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105637459 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002311_300 |
0.3084621734 |
- |
- |
PREDICTED: J protein JJJ2 [Jatropha curcas] |
| 18 |
Hb_002081_170 |
0.3085206817 |
- |
- |
Flavonol 3-sulfotransferase, putative [Ricinus communis] |
| 19 |
Hb_100743_010 |
0.3100056557 |
- |
- |
hypothetical protein JCGZ_17104 [Jatropha curcas] |
| 20 |
Hb_000109_230 |
0.3137136777 |
- |
- |
conserved hypothetical protein [Ricinus communis] |