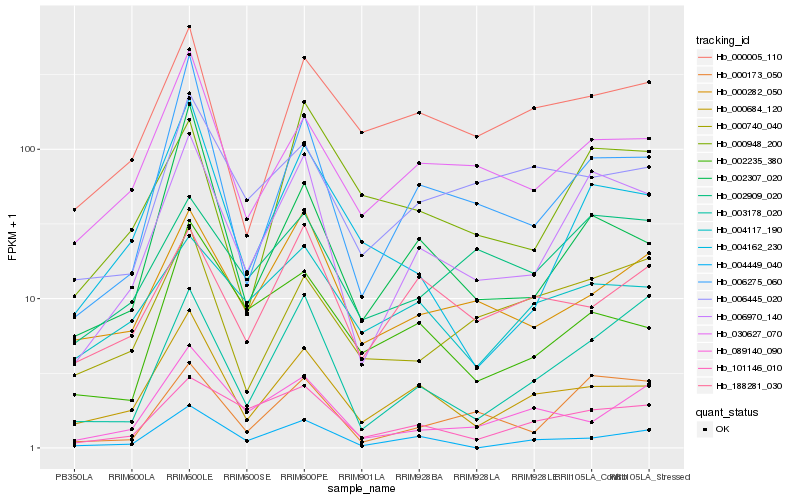
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006970_140 |
0.0 |
- |
- |
PREDICTED: histone H3.2-like [Solanum lycopersicum] |
| 2 |
Hb_000173_050 |
0.1289570833 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 3 |
Hb_003178_020 |
0.1658819996 |
transcription factor |
TF Family: B3 |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_006275_060 |
0.1700052926 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 5 |
Hb_101146_010 |
0.1746178195 |
- |
- |
PREDICTED: peregrin-like [Jatropha curcas] |
| 6 |
Hb_000684_120 |
0.1855966805 |
- |
- |
PREDICTED: uncharacterized protein LOC105642190 isoform X2 [Jatropha curcas] |
| 7 |
Hb_030627_070 |
0.1856021868 |
- |
- |
histone H4 [Zea mays] |
| 8 |
Hb_000282_050 |
0.1877486499 |
- |
- |
calcineurin B, putative [Ricinus communis] |
| 9 |
Hb_002909_020 |
0.1894425143 |
- |
- |
PREDICTED: uncharacterized protein LOC105632476 [Jatropha curcas] |
| 10 |
Hb_000948_200 |
0.1942744066 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 11 |
Hb_004162_230 |
0.1996294515 |
- |
- |
PREDICTED: histone H2B [Jatropha curcas] |
| 12 |
Hb_004449_040 |
0.202450302 |
- |
- |
PREDICTED: uncharacterized protein C630.12 [Jatropha curcas] |
| 13 |
Hb_000005_110 |
0.2084489054 |
- |
- |
histone H2B1 [Hevea brasiliensis] |
| 14 |
Hb_000740_040 |
0.2107303532 |
- |
- |
PREDICTED: uncharacterized protein LOC105126082 [Populus euphratica] |
| 15 |
Hb_188281_030 |
0.212412776 |
- |
- |
PREDICTED: uridine nucleosidase 1 [Jatropha curcas] |
| 16 |
Hb_004117_190 |
0.2128914628 |
- |
- |
PREDICTED: uncharacterized protein LOC105649109 [Jatropha curcas] |
| 17 |
Hb_002235_380 |
0.2137879249 |
- |
- |
hypothetical protein JCGZ_19860 [Jatropha curcas] |
| 18 |
Hb_002307_020 |
0.2145707299 |
- |
- |
PREDICTED: histone H1-like [Jatropha curcas] |
| 19 |
Hb_089140_090 |
0.2165152743 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like isoform X3 [Beta vulgaris subsp. vulgaris] |
| 20 |
Hb_006445_020 |
0.2165884537 |
- |
- |
hypothetical protein L484_026741 [Morus notabilis] |