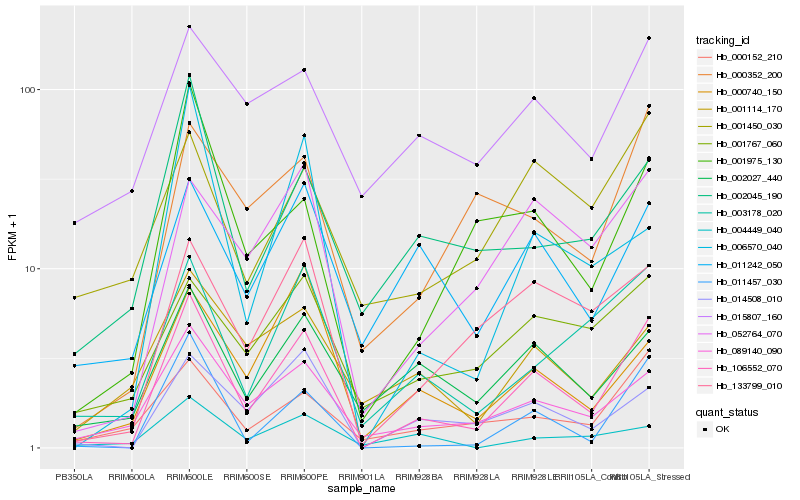
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_106552_070 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_011457_030 |
0.1676809999 |
- |
- |
type I inositol polyphosphate 5-phosphatase, putative [Ricinus communis] |
| 3 |
Hb_089140_090 |
0.1850191493 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like isoform X3 [Beta vulgaris subsp. vulgaris] |
| 4 |
Hb_014508_010 |
0.1914005874 |
- |
- |
PREDICTED: uncharacterized protein LOC105629998 [Jatropha curcas] |
| 5 |
Hb_003178_020 |
0.1918578319 |
transcription factor |
TF Family: B3 |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_133799_010 |
0.1980501032 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_052764_070 |
0.2034047501 |
- |
- |
PREDICTED: uncharacterized protein LOC105649473 [Jatropha curcas] |
| 8 |
Hb_001975_130 |
0.2038033795 |
- |
- |
PREDICTED: probable galactinol--sucrose galactosyltransferase 6 isoform X1 [Populus euphratica] |
| 9 |
Hb_000152_210 |
0.2055387208 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002027_440 |
0.2115157499 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_011242_050 |
0.2189454991 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2-23 kDa [Jatropha curcas] |
| 12 |
Hb_002045_190 |
0.2190381723 |
- |
- |
PREDICTED: histone H2A variant 1 [Jatropha curcas] |
| 13 |
Hb_000352_200 |
0.2225188685 |
- |
- |
PREDICTED: OTU domain-containing protein DDB_G0284757 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000740_150 |
0.2261563644 |
- |
- |
PREDICTED: uncharacterized protein LOC105629998 [Jatropha curcas] |
| 15 |
Hb_001767_060 |
0.2317462694 |
- |
- |
PREDICTED: sulfate transporter 4.1, chloroplastic-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_006570_040 |
0.2327013786 |
- |
- |
hypothetical protein CICLE_v10002683mg [Citrus clementina] |
| 17 |
Hb_004449_040 |
0.2352858698 |
- |
- |
PREDICTED: uncharacterized protein C630.12 [Jatropha curcas] |
| 18 |
Hb_001450_030 |
0.2368841379 |
- |
- |
PREDICTED: uncharacterized protein LOC105650827 [Jatropha curcas] |
| 19 |
Hb_015807_160 |
0.2396340381 |
- |
- |
PREDICTED: probable histone H2A.1 [Jatropha curcas] |
| 20 |
Hb_001114_170 |
0.2408064392 |
- |
- |
conserved hypothetical protein [Ricinus communis] |