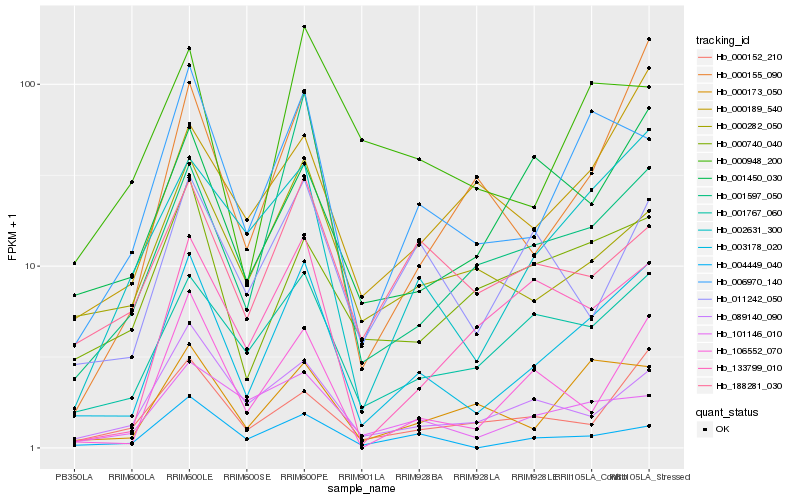
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003178_020 |
0.0 |
transcription factor |
TF Family: B3 |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_006970_140 |
0.1658819996 |
- |
- |
PREDICTED: histone H3.2-like [Solanum lycopersicum] |
| 3 |
Hb_002631_300 |
0.1666090864 |
- |
- |
PREDICTED: uncharacterized protein LOC105646181 [Jatropha curcas] |
| 4 |
Hb_001767_060 |
0.1749610362 |
- |
- |
PREDICTED: sulfate transporter 4.1, chloroplastic-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_000173_050 |
0.1832668826 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 6 |
Hb_106552_070 |
0.1918578319 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_004449_040 |
0.1944168327 |
- |
- |
PREDICTED: uncharacterized protein C630.12 [Jatropha curcas] |
| 8 |
Hb_000152_210 |
0.1983226625 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000282_050 |
0.199659034 |
- |
- |
calcineurin B, putative [Ricinus communis] |
| 10 |
Hb_089140_090 |
0.2036539477 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like isoform X3 [Beta vulgaris subsp. vulgaris] |
| 11 |
Hb_000155_090 |
0.2039651244 |
- |
- |
PREDICTED: glutaredoxin-C9 [Jatropha curcas] |
| 12 |
Hb_101146_010 |
0.204669423 |
- |
- |
PREDICTED: peregrin-like [Jatropha curcas] |
| 13 |
Hb_001597_050 |
0.211892719 |
- |
- |
- |
| 14 |
Hb_000740_040 |
0.2118997698 |
- |
- |
PREDICTED: uncharacterized protein LOC105126082 [Populus euphratica] |
| 15 |
Hb_011242_050 |
0.2151926352 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2-23 kDa [Jatropha curcas] |
| 16 |
Hb_000948_200 |
0.2161869715 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 17 |
Hb_133799_010 |
0.2165674039 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001450_030 |
0.2170218048 |
- |
- |
PREDICTED: uncharacterized protein LOC105650827 [Jatropha curcas] |
| 19 |
Hb_188281_030 |
0.217593281 |
- |
- |
PREDICTED: uridine nucleosidase 1 [Jatropha curcas] |
| 20 |
Hb_000189_540 |
0.2181923641 |
- |
- |
PREDICTED: tRNA(adenine(34)) deaminase, chloroplastic [Jatropha curcas] |