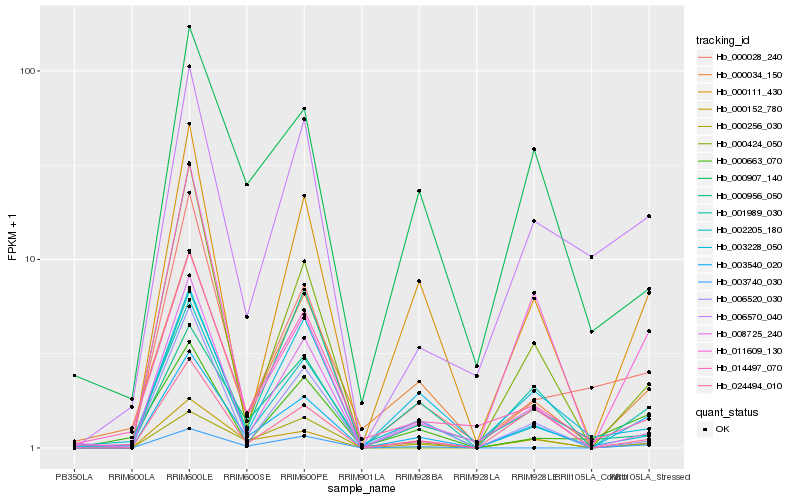
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003540_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000152_780 |
0.1417654184 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 3 |
Hb_000111_430 |
0.1598921996 |
- |
- |
PREDICTED: cytosolic sulfotransferase 15-like [Jatropha curcas] |
| 4 |
Hb_024494_010 |
0.1894447103 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 5 |
Hb_006570_040 |
0.1949599723 |
- |
- |
hypothetical protein CICLE_v10002683mg [Citrus clementina] |
| 6 |
Hb_000034_150 |
0.1950554734 |
- |
- |
PREDICTED: kinesin-like protein BC2 [Jatropha curcas] |
| 7 |
Hb_008725_240 |
0.2078502254 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000663_070 |
0.2107811122 |
- |
- |
cyclin A, putative [Ricinus communis] |
| 9 |
Hb_001989_030 |
0.2149251032 |
- |
- |
PREDICTED: copper transport protein ATX1 [Jatropha curcas] |
| 10 |
Hb_000028_240 |
0.2172314914 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000956_050 |
0.2182282798 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor SPATULA-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_002205_180 |
0.218677993 |
- |
- |
PREDICTED: uncharacterized protein LOC105649178 [Jatropha curcas] |
| 13 |
Hb_003740_030 |
0.2202024101 |
transcription factor |
TF Family: RAV |
DNA-binding protein RAV1, putative [Ricinus communis] |
| 14 |
Hb_011609_130 |
0.2250074779 |
- |
- |
- |
| 15 |
Hb_000424_050 |
0.2254902567 |
- |
- |
erecta, putative [Ricinus communis] |
| 16 |
Hb_000256_030 |
0.2257653414 |
- |
- |
transferase, putative [Ricinus communis] |
| 17 |
Hb_006520_030 |
0.2265577655 |
- |
- |
PREDICTED: osmotic avoidance abnormal protein 3 [Jatropha curcas] |
| 18 |
Hb_003228_050 |
0.227926395 |
- |
- |
chromomethylase [Hevea brasiliensis] |
| 19 |
Hb_000907_140 |
0.2281316102 |
- |
- |
PREDICTED: annexin D5-like [Jatropha curcas] |
| 20 |
Hb_014497_070 |
0.2286899714 |
- |
- |
PREDICTED: cyclin-dependent kinase B2-2 [Jatropha curcas] |