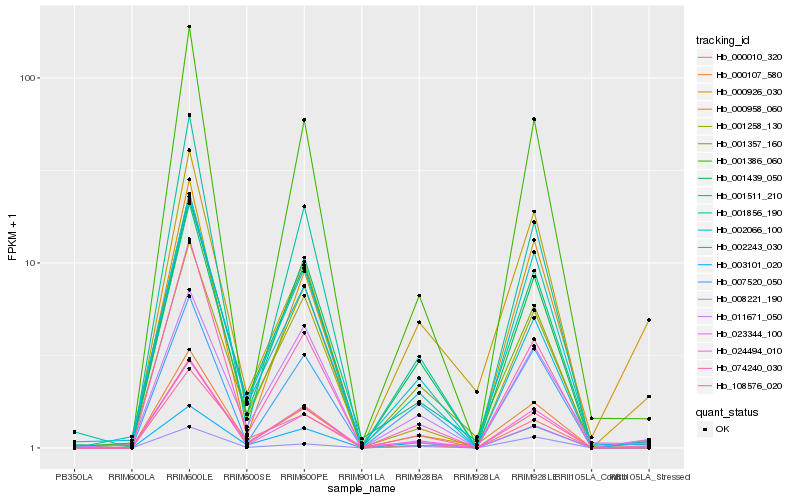
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_024494_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 2 |
Hb_007520_050 |
0.1312378996 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor WER-like [Jatropha curcas] |
| 3 |
Hb_001357_160 |
0.1324620928 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000107_580 |
0.1370275372 |
- |
- |
PREDICTED: delta(24)-sterol reductase [Jatropha curcas] |
| 5 |
Hb_001386_060 |
0.1388039326 |
- |
- |
PREDICTED: acetolactate synthase 1, chloroplastic-like [Vitis vinifera] |
| 6 |
Hb_001856_190 |
0.1407024498 |
- |
- |
SP1L, putative [Ricinus communis] |
| 7 |
Hb_001439_050 |
0.1430863398 |
- |
- |
plastocyanin-like domain-containing family protein [Populus trichocarpa] |
| 8 |
Hb_002066_100 |
0.14423913 |
- |
- |
PREDICTED: protein disulfide-isomerase SCO2 [Jatropha curcas] |
| 9 |
Hb_074240_030 |
0.1444168648 |
transcription factor |
TF Family: HMG |
PREDICTED: high mobility group B protein 3-like [Jatropha curcas] |
| 10 |
Hb_001511_210 |
0.1449645229 |
- |
- |
PREDICTED: protein ESKIMO 1-like [Jatropha curcas] |
| 11 |
Hb_008221_190 |
0.1453754603 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 [Jatropha curcas] |
| 12 |
Hb_002243_030 |
0.1455014375 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 3-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_011671_050 |
0.1497102219 |
- |
- |
PREDICTED: F-box protein At5g07670 [Jatropha curcas] |
| 14 |
Hb_000958_060 |
0.1510747838 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000010_320 |
0.1511869162 |
- |
- |
PREDICTED: protein STICHEL-like 2 [Jatropha curcas] |
| 16 |
Hb_003101_020 |
0.1513657677 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor NAM-B2 [Jatropha curcas] |
| 17 |
Hb_001258_130 |
0.1527425226 |
- |
- |
PREDICTED: kinesin-like protein KIF19 [Jatropha curcas] |
| 18 |
Hb_108576_020 |
0.1539024104 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_023344_100 |
0.1553299662 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.2 [Jatropha curcas] |
| 20 |
Hb_000926_030 |
0.1553332134 |
- |
- |
zinc finger protein, putative [Ricinus communis] |