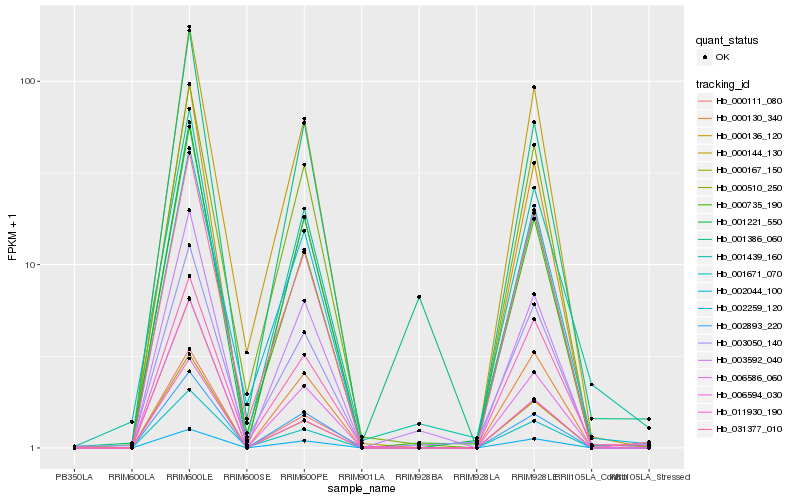
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001439_160 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644097 [Jatropha curcas] |
| 2 |
Hb_003050_140 |
0.0816720815 |
- |
- |
PREDICTED: potassium channel KAT1 [Jatropha curcas] |
| 3 |
Hb_001221_550 |
0.1000813344 |
- |
- |
Corticosteroid 11-beta-dehydrogenase, putative [Ricinus communis] |
| 4 |
Hb_011930_190 |
0.1002069914 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 17 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001386_060 |
0.1018276902 |
- |
- |
PREDICTED: acetolactate synthase 1, chloroplastic-like [Vitis vinifera] |
| 6 |
Hb_003592_040 |
0.1067136781 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 9 [Jatropha curcas] |
| 7 |
Hb_000735_190 |
0.1099728521 |
- |
- |
hypothetical protein POPTR_0006s19340g [Populus trichocarpa] |
| 8 |
Hb_000130_340 |
0.1100224447 |
- |
- |
- |
| 9 |
Hb_002044_100 |
0.1108911302 |
- |
- |
PREDICTED: uncharacterized protein LOC105643555 [Jatropha curcas] |
| 10 |
Hb_001671_070 |
0.1124681904 |
- |
- |
kinase, putative [Ricinus communis] |
| 11 |
Hb_000144_130 |
0.1129321621 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000510_250 |
0.1139328092 |
transcription factor |
TF Family: AUX/IAA |
auxin-induced protein aux28 [Populus trichocarpa] |
| 13 |
Hb_031377_010 |
0.1146203716 |
transcription factor |
TF Family: C2C2-YABBY |
unknown [Populus trichocarpa] |
| 14 |
Hb_002893_220 |
0.1164350495 |
- |
- |
hypothetical protein B456_001G004300 [Gossypium raimondii] |
| 15 |
Hb_006586_060 |
0.1179905979 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 16 |
Hb_002259_120 |
0.118716193 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 17 |
Hb_006594_030 |
0.1204963396 |
- |
- |
- |
| 18 |
Hb_000111_080 |
0.1210161933 |
- |
- |
hypothetical protein JCGZ_24534 [Jatropha curcas] |
| 19 |
Hb_000136_120 |
0.1215056277 |
- |
- |
PREDICTED: probable strigolactone esterase D14 homolog [Jatropha curcas] |
| 20 |
Hb_000167_150 |
0.1228085682 |
- |
- |
hypothetical protein POPTR_0008s03590g [Populus trichocarpa] |