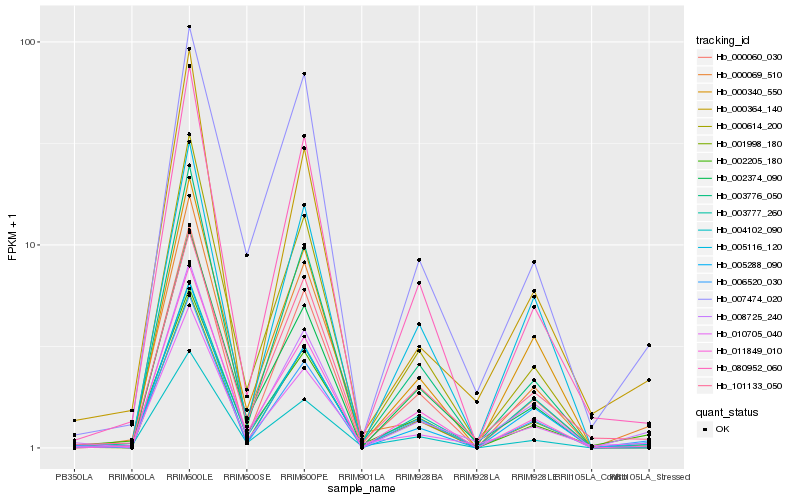
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008725_240 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_006520_030 |
0.0988066744 |
- |
- |
PREDICTED: osmotic avoidance abnormal protein 3 [Jatropha curcas] |
| 3 |
Hb_010705_040 |
0.1053371365 |
- |
- |
PREDICTED: kinesin-4 [Jatropha curcas] |
| 4 |
Hb_005288_090 |
0.1141319257 |
- |
- |
PREDICTED: uncharacterized protein LOC105647753 [Jatropha curcas] |
| 5 |
Hb_003777_260 |
0.1163755961 |
- |
- |
PREDICTED: uncharacterized protein LOC105640927 isoform X4 [Jatropha curcas] |
| 6 |
Hb_000364_140 |
0.117139157 |
- |
- |
PREDICTED: uncharacterized acetyltransferase At3g50280-like [Jatropha curcas] |
| 7 |
Hb_002374_090 |
0.119582222 |
- |
- |
PREDICTED: cyclin-A1-1 [Jatropha curcas] |
| 8 |
Hb_001998_180 |
0.1197407743 |
- |
- |
actin binding protein, putative [Ricinus communis] |
| 9 |
Hb_004102_090 |
0.1249308629 |
transcription factor |
TF Family: CPP |
hypothetical protein JCGZ_06186 [Jatropha curcas] |
| 10 |
Hb_011849_010 |
0.1252229689 |
- |
- |
PREDICTED: kinesin-like protein KIFC3 isoform X4 [Jatropha curcas] |
| 11 |
Hb_003776_050 |
0.1262957303 |
- |
- |
PREDICTED: uncharacterized protein LOC105641219 [Jatropha curcas] |
| 12 |
Hb_000614_200 |
0.1264344769 |
- |
- |
plant mitotic spindle assembly checkpoint protein mad2, putative [Ricinus communis] |
| 13 |
Hb_007474_020 |
0.1268470955 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 14 |
Hb_101133_050 |
0.1277918274 |
- |
- |
PREDICTED: condensin complex subunit 3 [Jatropha curcas] |
| 15 |
Hb_080952_060 |
0.1278235547 |
- |
- |
PREDICTED: patellin-4 [Populus euphratica] |
| 16 |
Hb_002205_180 |
0.1278356168 |
- |
- |
PREDICTED: uncharacterized protein LOC105649178 [Jatropha curcas] |
| 17 |
Hb_000340_550 |
0.1280073608 |
- |
- |
PREDICTED: rho GTPase-activating protein 2 [Jatropha curcas] |
| 18 |
Hb_000069_510 |
0.1281577798 |
- |
- |
PREDICTED: histone H3-like centromeric protein HTR12 [Jatropha curcas] |
| 19 |
Hb_005116_120 |
0.1294138238 |
- |
- |
PREDICTED: uncharacterized protein LOC105649997 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000060_030 |
0.1296282772 |
- |
- |
PREDICTED: carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1-like [Jatropha curcas] |