| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000638_240 |
0.0 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0234100-like [Prunus mume] |
| 2 |
Hb_000260_380 |
0.1357196798 |
- |
- |
PREDICTED: cell division control protein 2 homolog C [Jatropha curcas] |
| 3 |
Hb_005542_150 |
0.1364235445 |
- |
- |
PREDICTED: uncharacterized protein LOC105649178 [Jatropha curcas] |
| 4 |
Hb_000576_140 |
0.1405489737 |
- |
- |
PREDICTED: uncharacterized protein LOC105638282 [Jatropha curcas] |
| 5 |
Hb_001999_160 |
0.1407382116 |
- |
- |
hypothetical protein JCGZ_08488 [Jatropha curcas] |
| 6 |
Hb_003109_010 |
0.1417185863 |
- |
- |
PREDICTED: kinesin-like protein KIN12B [Jatropha curcas] |
| 7 |
Hb_003106_090 |
0.1420977692 |
transcription factor |
TF Family: E2F-DP |
E2F, putative [Ricinus communis] |
| 8 |
Hb_003813_090 |
0.1421282023 |
- |
- |
PREDICTED: uncharacterized protein LOC105638743 [Jatropha curcas] |
| 9 |
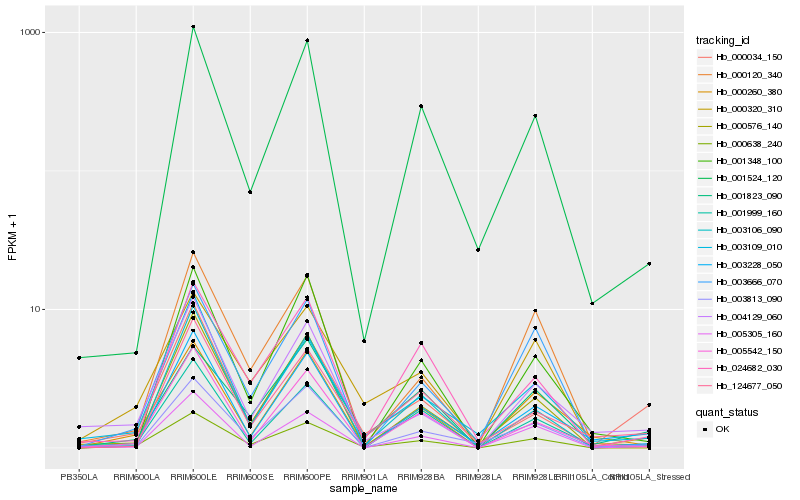
Hb_001524_120 |
0.1460859367 |
- |
- |
myo-inositol-1 phosphate synthase [Hevea brasiliensis] |
| 10 |
Hb_003228_050 |
0.1472366744 |
- |
- |
chromomethylase [Hevea brasiliensis] |
| 11 |
Hb_005305_160 |
0.1480050725 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_024682_030 |
0.1481524391 |
- |
- |
PREDICTED: DNA replication licensing factor MCM2 [Jatropha curcas] |
| 13 |
Hb_004129_060 |
0.1484514028 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 14 |
Hb_001823_090 |
0.1496875727 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_003666_070 |
0.1497188167 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g66560 [Jatropha curcas] |
| 16 |
Hb_000320_310 |
0.1498910217 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 13C [Jatropha curcas] |
| 17 |
Hb_000120_340 |
0.1518288233 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 18 |
Hb_124677_050 |
0.1535319512 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000034_150 |
0.1536458688 |
- |
- |
PREDICTED: kinesin-like protein BC2 [Jatropha curcas] |
| 20 |
Hb_001348_100 |
0.1540411495 |
- |
- |
PREDICTED: uncharacterized protein LOC105638934 [Jatropha curcas] |