| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
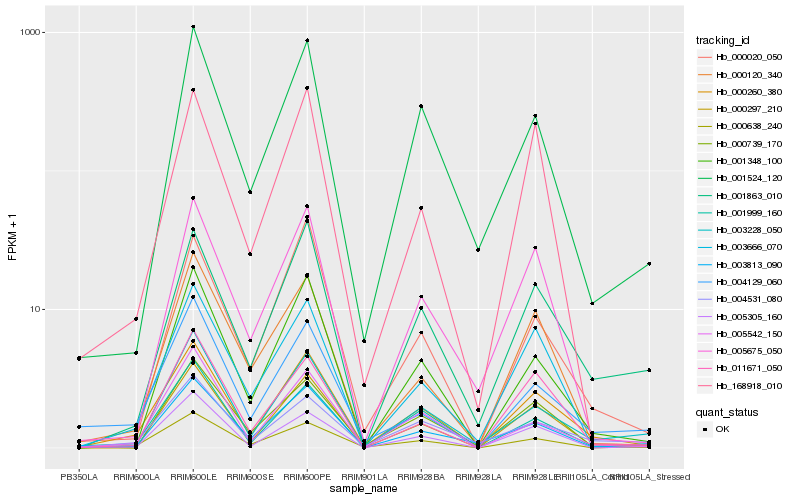
Hb_000260_380 |
0.0 |
- |
- |
PREDICTED: cell division control protein 2 homolog C [Jatropha curcas] |
| 2 |
Hb_000297_210 |
0.1252981748 |
- |
- |
Bipolar kinesin KRP-130, putative [Ricinus communis] |
| 3 |
Hb_005542_150 |
0.1291194828 |
- |
- |
PREDICTED: uncharacterized protein LOC105649178 [Jatropha curcas] |
| 4 |
Hb_001348_100 |
0.1320112794 |
- |
- |
PREDICTED: uncharacterized protein LOC105638934 [Jatropha curcas] |
| 5 |
Hb_001999_160 |
0.1332208875 |
- |
- |
hypothetical protein JCGZ_08488 [Jatropha curcas] |
| 6 |
Hb_003666_070 |
0.1343820554 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g66560 [Jatropha curcas] |
| 7 |
Hb_000638_240 |
0.1357196798 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0234100-like [Prunus mume] |
| 8 |
Hb_001863_010 |
0.1372363872 |
- |
- |
PREDICTED: classical arabinogalactan protein 4-like [Jatropha curcas] |
| 9 |
Hb_004531_080 |
0.1397300496 |
- |
- |
hypothetical protein RCOM_1491960 [Ricinus communis] |
| 10 |
Hb_001524_120 |
0.1406886946 |
- |
- |
myo-inositol-1 phosphate synthase [Hevea brasiliensis] |
| 11 |
Hb_003813_090 |
0.14445631 |
- |
- |
PREDICTED: uncharacterized protein LOC105638743 [Jatropha curcas] |
| 12 |
Hb_003228_050 |
0.1444940568 |
- |
- |
chromomethylase [Hevea brasiliensis] |
| 13 |
Hb_000120_340 |
0.1468375916 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 14 |
Hb_000739_170 |
0.1493561433 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 4 [Jatropha curcas] |
| 15 |
Hb_011671_050 |
0.1494435648 |
- |
- |
PREDICTED: F-box protein At5g07670 [Jatropha curcas] |
| 16 |
Hb_005305_160 |
0.149635978 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_005675_050 |
0.1502693954 |
- |
- |
alpha-glucosidase, putative [Ricinus communis] |
| 18 |
Hb_004129_060 |
0.1518355395 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 19 |
Hb_168918_010 |
0.1520192176 |
- |
- |
pectin acetylesterase, putative [Ricinus communis] |
| 20 |
Hb_000020_050 |
0.1529076462 |
- |
- |
PREDICTED: expansin-A6 [Jatropha curcas] |