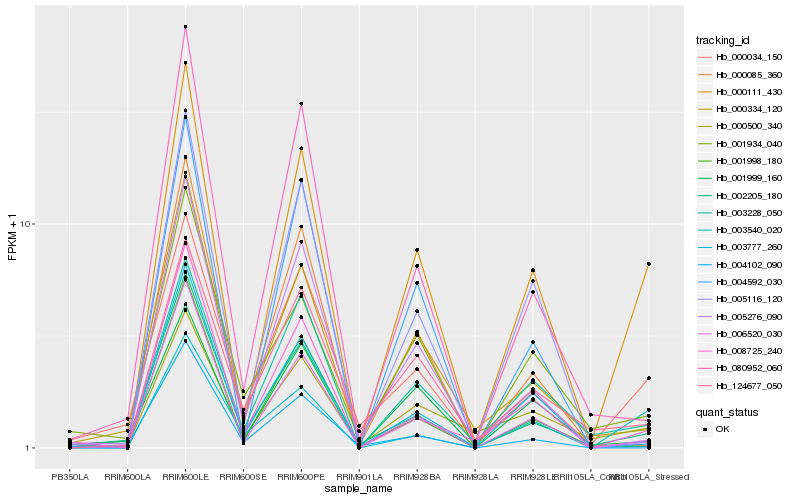
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000111_430 |
0.0 |
- |
- |
PREDICTED: cytosolic sulfotransferase 15-like [Jatropha curcas] |
| 2 |
Hb_006520_030 |
0.1384528112 |
- |
- |
PREDICTED: osmotic avoidance abnormal protein 3 [Jatropha curcas] |
| 3 |
Hb_008725_240 |
0.1549092136 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_003540_020 |
0.1598921996 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000034_150 |
0.1624085543 |
- |
- |
PREDICTED: kinesin-like protein BC2 [Jatropha curcas] |
| 6 |
Hb_003228_050 |
0.1627322524 |
- |
- |
chromomethylase [Hevea brasiliensis] |
| 7 |
Hb_002205_180 |
0.1669428405 |
- |
- |
PREDICTED: uncharacterized protein LOC105649178 [Jatropha curcas] |
| 8 |
Hb_000500_340 |
0.175995779 |
- |
- |
PREDICTED: exonuclease 1 [Jatropha curcas] |
| 9 |
Hb_000334_120 |
0.1785032061 |
- |
- |
PREDICTED: DNA topoisomerase 2 [Jatropha curcas] |
| 10 |
Hb_005116_120 |
0.181571158 |
- |
- |
PREDICTED: uncharacterized protein LOC105649997 isoform X2 [Jatropha curcas] |
| 11 |
Hb_001998_180 |
0.1841154785 |
- |
- |
actin binding protein, putative [Ricinus communis] |
| 12 |
Hb_004102_090 |
0.1842391299 |
transcription factor |
TF Family: CPP |
hypothetical protein JCGZ_06186 [Jatropha curcas] |
| 13 |
Hb_005276_090 |
0.186807236 |
- |
- |
PREDICTED: uncharacterized protein LOC105649997 isoform X2 [Jatropha curcas] |
| 14 |
Hb_080952_060 |
0.1872637529 |
- |
- |
PREDICTED: patellin-4 [Populus euphratica] |
| 15 |
Hb_003777_260 |
0.1874517928 |
- |
- |
PREDICTED: uncharacterized protein LOC105640927 isoform X4 [Jatropha curcas] |
| 16 |
Hb_001934_040 |
0.1891952312 |
- |
- |
PREDICTED: vacuolar iron transporter 1-like [Jatropha curcas] |
| 17 |
Hb_124677_050 |
0.1893751584 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000085_360 |
0.1901096883 |
- |
- |
PREDICTED: kinesin-like protein KIN12B [Jatropha curcas] |
| 19 |
Hb_004592_030 |
0.1919550027 |
- |
- |
unknown [Lotus japonicus] |
| 20 |
Hb_001999_160 |
0.1929515896 |
- |
- |
hypothetical protein JCGZ_08488 [Jatropha curcas] |