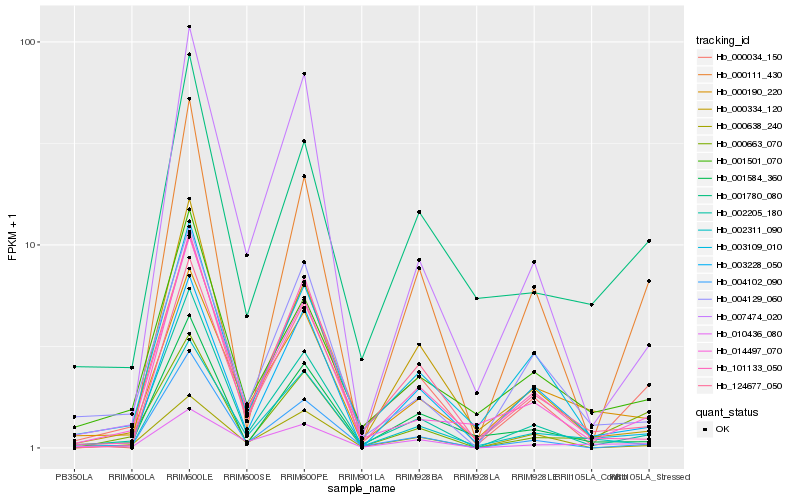
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000034_150 |
0.0 |
- |
- |
PREDICTED: kinesin-like protein BC2 [Jatropha curcas] |
| 2 |
Hb_002205_180 |
0.1177085993 |
- |
- |
PREDICTED: uncharacterized protein LOC105649178 [Jatropha curcas] |
| 3 |
Hb_014497_070 |
0.1423579463 |
- |
- |
PREDICTED: cyclin-dependent kinase B2-2 [Jatropha curcas] |
| 4 |
Hb_004102_090 |
0.143545361 |
transcription factor |
TF Family: CPP |
hypothetical protein JCGZ_06186 [Jatropha curcas] |
| 5 |
Hb_124677_050 |
0.1451586132 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000663_070 |
0.1524975163 |
- |
- |
cyclin A, putative [Ricinus communis] |
| 7 |
Hb_000638_240 |
0.1536458688 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0234100-like [Prunus mume] |
| 8 |
Hb_101133_050 |
0.1583457519 |
- |
- |
PREDICTED: condensin complex subunit 3 [Jatropha curcas] |
| 9 |
Hb_002311_090 |
0.1601047455 |
- |
- |
PREDICTED: uncharacterized protein LOC105643503 [Jatropha curcas] |
| 10 |
Hb_003228_050 |
0.1607693315 |
- |
- |
chromomethylase [Hevea brasiliensis] |
| 11 |
Hb_003109_010 |
0.1611389757 |
- |
- |
PREDICTED: kinesin-like protein KIN12B [Jatropha curcas] |
| 12 |
Hb_010436_080 |
0.1617618817 |
- |
- |
PREDICTED: DNA replication licensing factor MCM5 [Jatropha curcas] |
| 13 |
Hb_000111_430 |
0.1624085543 |
- |
- |
PREDICTED: cytosolic sulfotransferase 15-like [Jatropha curcas] |
| 14 |
Hb_001501_070 |
0.1643579977 |
- |
- |
PREDICTED: condensin complex subunit 2 [Jatropha curcas] |
| 15 |
Hb_000190_220 |
0.1650524069 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek2 [Jatropha curcas] |
| 16 |
Hb_004129_060 |
0.1650593118 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 17 |
Hb_001780_080 |
0.1657022644 |
- |
- |
PREDICTED: tubulin beta-2 chain [Jatropha curcas] |
| 18 |
Hb_007474_020 |
0.1657884374 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 19 |
Hb_001584_360 |
0.16580521 |
- |
- |
PREDICTED: serine/threonine-protein kinase haspin homolog hrk1 [Jatropha curcas] |
| 20 |
Hb_000334_120 |
0.1677177064 |
- |
- |
PREDICTED: DNA topoisomerase 2 [Jatropha curcas] |