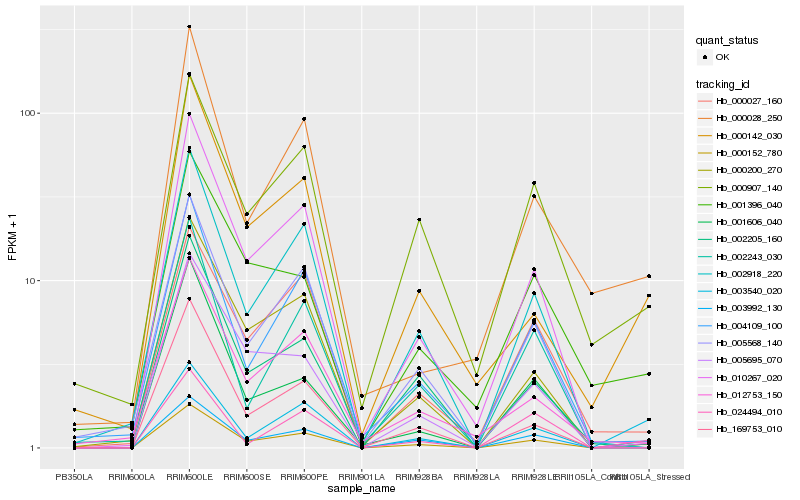
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000152_780 |
0.0 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 2 |
Hb_003540_020 |
0.1417654184 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_010267_020 |
0.1524846036 |
- |
- |
diphosphoinositol polyphosphate phosphohydrolase, putative [Ricinus communis] |
| 4 |
Hb_000142_030 |
0.1535534809 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 5 |
Hb_000907_140 |
0.1539284093 |
- |
- |
PREDICTED: annexin D5-like [Jatropha curcas] |
| 6 |
Hb_003992_130 |
0.162951334 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 7 |
Hb_002918_220 |
0.1647305897 |
- |
- |
PREDICTED: uncharacterized protein LOC105138896 [Populus euphratica] |
| 8 |
Hb_004109_100 |
0.1680895834 |
- |
- |
Interactor of constitutive active rops 1 isoform 1 [Theobroma cacao] |
| 9 |
Hb_024494_010 |
0.1690104359 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 10 |
Hb_005568_140 |
0.1705203734 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 11 |
Hb_000027_160 |
0.1717721957 |
- |
- |
PREDICTED: probable protein phosphatase 2C 15 isoform X3 [Jatropha curcas] |
| 12 |
Hb_001396_040 |
0.1733120171 |
- |
- |
PREDICTED: uncharacterized protein LOC105644845 [Jatropha curcas] |
| 13 |
Hb_000028_250 |
0.1736558667 |
- |
- |
PREDICTED: nudix hydrolase 4 [Jatropha curcas] |
| 14 |
Hb_002205_160 |
0.1740502321 |
- |
- |
chalcone isomerase [Hibiscus cannabinus] |
| 15 |
Hb_000200_270 |
0.1743992372 |
- |
- |
PREDICTED: lipid phosphate phosphatase 1-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_002243_030 |
0.1755659205 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 3-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_169753_010 |
0.1758343458 |
- |
- |
hypothetical protein JCGZ_18439 [Jatropha curcas] |
| 18 |
Hb_001606_040 |
0.1761858748 |
- |
- |
Rho GDP-dissociation inhibitor, putative [Ricinus communis] |
| 19 |
Hb_005695_070 |
0.1772534112 |
- |
- |
Cysteine-rich RLK 29 [Theobroma cacao] |
| 20 |
Hb_012753_150 |
0.1779229948 |
transcription factor |
TF Family: GRAS |
DELLA protein RGL1, putative [Ricinus communis] |