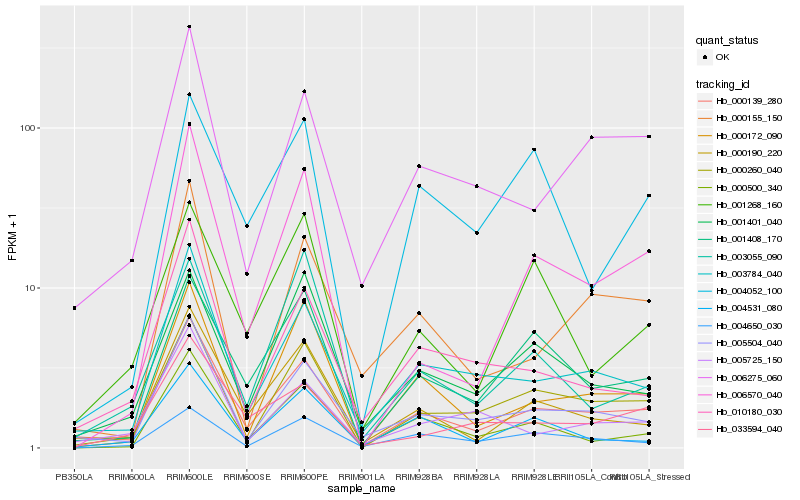
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000260_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649093 [Jatropha curcas] |
| 2 |
Hb_000139_280 |
0.1349767281 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase NPK1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_005504_040 |
0.1427889408 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_004650_030 |
0.1431495678 |
- |
- |
PREDICTED: uncharacterized protein LOC105638888 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001401_040 |
0.1479005768 |
- |
- |
PREDICTED: protein CYPRO4 [Jatropha curcas] |
| 6 |
Hb_003784_040 |
0.1526677229 |
- |
- |
Afadin- and alpha-actinin-binding protein, putative [Ricinus communis] |
| 7 |
Hb_000172_090 |
0.1551127066 |
- |
- |
PREDICTED: uncharacterized protein LOC105650755 [Jatropha curcas] |
| 8 |
Hb_001408_170 |
0.156302542 |
- |
- |
PREDICTED: phosphoglycolate phosphatase 2 [Jatropha curcas] |
| 9 |
Hb_006275_060 |
0.1648579587 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 10 |
Hb_006570_040 |
0.1806825739 |
- |
- |
hypothetical protein CICLE_v10002683mg [Citrus clementina] |
| 11 |
Hb_000500_340 |
0.1874837226 |
- |
- |
PREDICTED: exonuclease 1 [Jatropha curcas] |
| 12 |
Hb_000155_150 |
0.1922591951 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 13 |
Hb_003055_090 |
0.1955730784 |
- |
- |
PREDICTED: uncharacterized protein LOC105649490 [Jatropha curcas] |
| 14 |
Hb_005725_150 |
0.1968331519 |
- |
- |
PREDICTED: kinesin-related protein 11 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000190_220 |
0.1992527099 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek2 [Jatropha curcas] |
| 16 |
Hb_010180_030 |
0.2009171492 |
- |
- |
PREDICTED: WEB family protein At2g38370-like [Jatropha curcas] |
| 17 |
Hb_004531_080 |
0.2014161923 |
- |
- |
hypothetical protein RCOM_1491960 [Ricinus communis] |
| 18 |
Hb_004052_100 |
0.2022773612 |
- |
- |
PREDICTED: protein SPIRAL1-like 3 [Jatropha curcas] |
| 19 |
Hb_001268_160 |
0.2023744897 |
- |
- |
Glycerophosphodiester phosphodiesterase [Medicago truncatula] |
| 20 |
Hb_033594_040 |
0.2024996513 |
- |
- |
PREDICTED: heptahelical transmembrane protein 4-like isoform X2 [Jatropha curcas] |