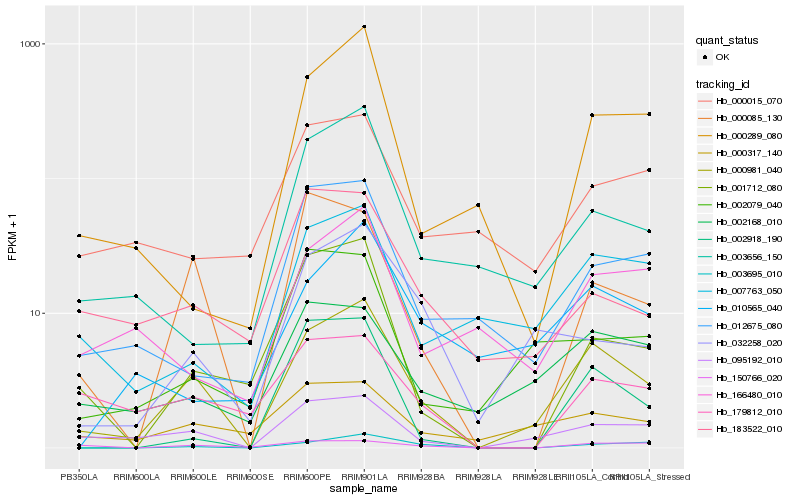
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000981_040 |
0.0 |
- |
- |
- |
| 2 |
Hb_095192_010 |
0.1901516821 |
- |
- |
hypothetical protein VITISV_043980 [Vitis vinifera] |
| 3 |
Hb_000085_130 |
0.234255554 |
- |
- |
- |
| 4 |
Hb_000317_140 |
0.2400192019 |
- |
- |
hypothetical protein CISIN_1g0019781mg, partial [Citrus sinensis] |
| 5 |
Hb_003695_010 |
0.240051516 |
- |
- |
PREDICTED: uncharacterized protein LOC104104926 [Nicotiana tomentosiformis] |
| 6 |
Hb_007763_050 |
0.2401476963 |
- |
- |
hypothetical protein L484_012752 [Morus notabilis] |
| 7 |
Hb_002168_010 |
0.241372579 |
- |
- |
PREDICTED: bifunctional dihydrofolate reductase-thymidylate synthase-like isoform X4 [Vitis vinifera] |
| 8 |
Hb_032258_020 |
0.2445420259 |
- |
- |
PREDICTED: amidase 1-like isoform X2 [Malus domestica] |
| 9 |
Hb_001712_080 |
0.2532916255 |
- |
- |
- |
| 10 |
Hb_003656_150 |
0.2557307705 |
- |
- |
- |
| 11 |
Hb_000289_080 |
0.2629171655 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_010565_040 |
0.2633281145 |
- |
- |
hypothetical protein POPTR_0015s05280g, partial [Populus trichocarpa] |
| 13 |
Hb_012675_080 |
0.2635388818 |
- |
- |
PREDICTED: cytoplasmic tRNA 2-thiolation protein 1 [Populus euphratica] |
| 14 |
Hb_002918_190 |
0.2648113909 |
- |
- |
Uncharacterized protein TCM_029816 [Theobroma cacao] |
| 15 |
Hb_002079_040 |
0.2658324001 |
- |
- |
hypothetical protein VITISV_018542 [Vitis vinifera] |
| 16 |
Hb_179812_010 |
0.2665871897 |
- |
- |
TdcA1-ORF1-ORF2 [Daucus carota] |
| 17 |
Hb_166480_010 |
0.2672543612 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase, mitochondrial-like [Cucumis melo] |
| 18 |
Hb_150766_020 |
0.272478866 |
- |
- |
hypothetical protein VITISV_030841 [Vitis vinifera] |
| 19 |
Hb_183522_010 |
0.2731023481 |
- |
- |
Far upstream element-binding protein, putative [Ricinus communis] |
| 20 |
Hb_000015_070 |
0.2745689586 |
- |
- |
hypothetical protein POPTR_0001s22690g [Populus trichocarpa] |