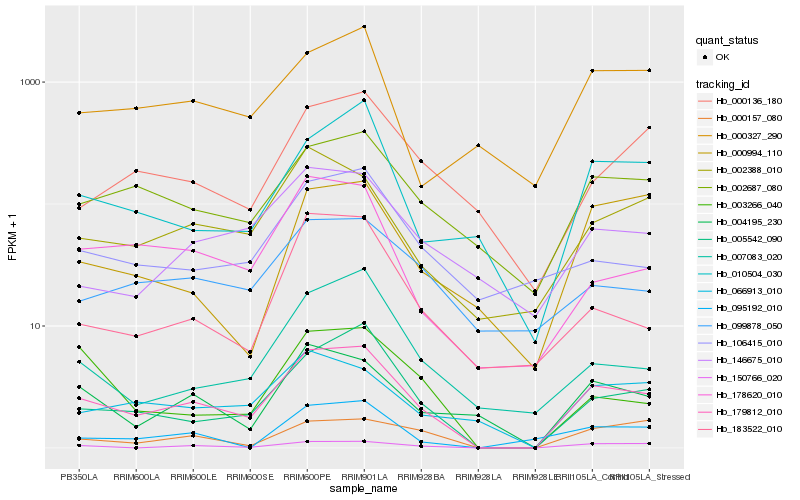
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_179812_010 |
0.0 |
- |
- |
TdcA1-ORF1-ORF2 [Daucus carota] |
| 2 |
Hb_005542_090 |
0.1511953624 |
- |
- |
PREDICTED: putative disease resistance protein RGA4, partial [Elaeis guineensis] |
| 3 |
Hb_004195_230 |
0.1794977871 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002388_010 |
0.179939128 |
- |
- |
- |
| 5 |
Hb_002687_080 |
0.1801935122 |
- |
- |
hypothetical protein JCGZ_06535 [Jatropha curcas] |
| 6 |
Hb_178620_010 |
0.1849469348 |
- |
- |
ADP-ribosylation factor, arf, putative [Ricinus communis] |
| 7 |
Hb_150766_020 |
0.1855783751 |
- |
- |
hypothetical protein VITISV_030841 [Vitis vinifera] |
| 8 |
Hb_010504_030 |
0.1903208449 |
- |
- |
calmodulin homolog {EST} [Brassica napus, Naehan, root, Peptide Partial, 53 aa] |
| 9 |
Hb_066913_010 |
0.1920482338 |
- |
- |
hypothetical protein POPTR_0006s03150g [Populus trichocarpa] |
| 10 |
Hb_003266_040 |
0.1993037001 |
- |
- |
PREDICTED: 6.7 kDa chloroplast outer envelope membrane protein-like [Jatropha curcas] |
| 11 |
Hb_000157_080 |
0.2008513275 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized N-acetyltransferase ycf52 [Jatropha curcas] |
| 12 |
Hb_007083_020 |
0.2052416065 |
- |
- |
hypothetical protein F775_15403 [Aegilops tauschii] |
| 13 |
Hb_183522_010 |
0.2072917331 |
- |
- |
Far upstream element-binding protein, putative [Ricinus communis] |
| 14 |
Hb_000327_290 |
0.2074780842 |
- |
- |
Histone H4 [Triticum urartu] |
| 15 |
Hb_000136_180 |
0.2088983313 |
- |
- |
ribosomal protein S28, putative [Ricinus communis] |
| 16 |
Hb_146675_010 |
0.2099176896 |
- |
- |
PREDICTED: peroxisomal nicotinamide adenine dinucleotide carrier-like isoform X2 [Malus domestica] |
| 17 |
Hb_099878_050 |
0.2104907614 |
- |
- |
PREDICTED: uncharacterized protein LOC105123495 isoform X2 [Populus euphratica] |
| 18 |
Hb_095192_010 |
0.2133072731 |
- |
- |
hypothetical protein VITISV_043980 [Vitis vinifera] |
| 19 |
Hb_106415_010 |
0.2142020863 |
- |
- |
PREDICTED: snRNA-activating protein complex subunit isoform X1 [Jatropha curcas] |
| 20 |
Hb_000994_110 |
0.2205962195 |
- |
- |
conserved hypothetical protein [Ricinus communis] |