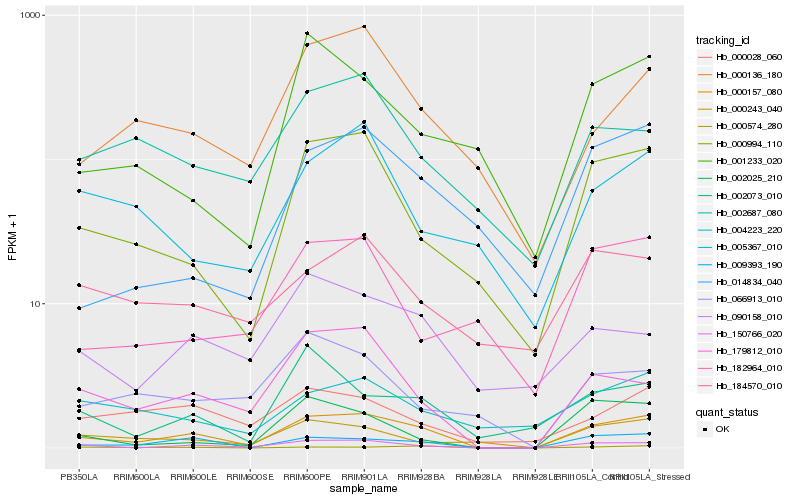
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000157_080 |
0.0 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized N-acetyltransferase ycf52 [Jatropha curcas] |
| 2 |
Hb_150766_020 |
0.1634430776 |
- |
- |
hypothetical protein VITISV_030841 [Vitis vinifera] |
| 3 |
Hb_009393_190 |
0.1711604902 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase AIP2-like [Jatropha curcas] |
| 4 |
Hb_000994_110 |
0.1900588732 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_179812_010 |
0.2008513275 |
- |
- |
TdcA1-ORF1-ORF2 [Daucus carota] |
| 6 |
Hb_000243_040 |
0.2240648667 |
- |
- |
- |
| 7 |
Hb_000136_180 |
0.2285774464 |
- |
- |
ribosomal protein S28, putative [Ricinus communis] |
| 8 |
Hb_000028_060 |
0.2355240803 |
- |
- |
- |
| 9 |
Hb_002073_010 |
0.2364156874 |
- |
- |
- |
| 10 |
Hb_002025_210 |
0.2365976773 |
- |
- |
- |
| 11 |
Hb_014834_040 |
0.2420651715 |
- |
- |
hypothetical protein JCGZ_03409 [Jatropha curcas] |
| 12 |
Hb_002687_080 |
0.2426693881 |
- |
- |
hypothetical protein JCGZ_06535 [Jatropha curcas] |
| 13 |
Hb_090158_010 |
0.2434935923 |
- |
- |
hypothetical protein RCOM_1968400 [Ricinus communis] |
| 14 |
Hb_000574_280 |
0.2451498257 |
- |
- |
hypothetical protein POPTR_0006s28780g [Populus trichocarpa] |
| 15 |
Hb_004223_220 |
0.2459537399 |
- |
- |
PREDICTED: uncharacterized protein LOC105635366 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001233_020 |
0.2479494589 |
- |
- |
hypothetical protein JCGZ_04109 [Jatropha curcas] |
| 17 |
Hb_005367_010 |
0.2479606672 |
- |
- |
- |
| 18 |
Hb_184570_010 |
0.2491180895 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 8 [Jatropha curcas] |
| 19 |
Hb_182964_010 |
0.2492182084 |
- |
- |
PREDICTED: uncharacterized protein LOC105642044 isoform X2 [Jatropha curcas] |
| 20 |
Hb_066913_010 |
0.2516943204 |
- |
- |
hypothetical protein POPTR_0006s03150g [Populus trichocarpa] |