| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
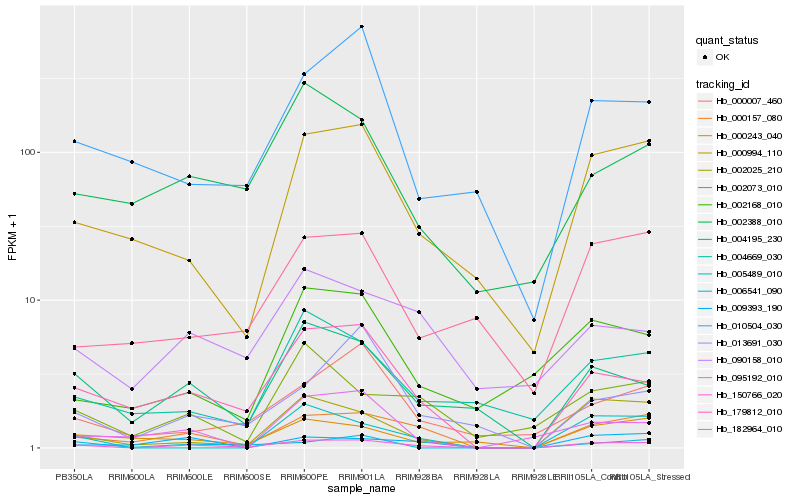
Hb_150766_020 |
0.0 |
- |
- |
hypothetical protein VITISV_030841 [Vitis vinifera] |
| 2 |
Hb_000157_080 |
0.1634430776 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized N-acetyltransferase ycf52 [Jatropha curcas] |
| 3 |
Hb_179812_010 |
0.1855783751 |
- |
- |
TdcA1-ORF1-ORF2 [Daucus carota] |
| 4 |
Hb_004195_230 |
0.2169233285 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002025_210 |
0.2219878147 |
- |
- |
- |
| 6 |
Hb_000994_110 |
0.2251266384 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_009393_190 |
0.2549855474 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase AIP2-like [Jatropha curcas] |
| 8 |
Hb_000243_040 |
0.255248991 |
- |
- |
- |
| 9 |
Hb_002073_010 |
0.2554710529 |
- |
- |
- |
| 10 |
Hb_090158_010 |
0.2571515043 |
- |
- |
hypothetical protein RCOM_1968400 [Ricinus communis] |
| 11 |
Hb_002388_010 |
0.2578249903 |
- |
- |
- |
| 12 |
Hb_006541_090 |
0.2592623881 |
- |
- |
hypothetical protein VITISV_020777 [Vitis vinifera] |
| 13 |
Hb_000007_460 |
0.2607135191 |
- |
- |
PREDICTED: uncharacterized protein LOC105641948 [Jatropha curcas] |
| 14 |
Hb_004669_030 |
0.2627815771 |
- |
- |
- |
| 15 |
Hb_010504_030 |
0.2652486992 |
- |
- |
calmodulin homolog {EST} [Brassica napus, Naehan, root, Peptide Partial, 53 aa] |
| 16 |
Hb_002168_010 |
0.265510419 |
- |
- |
PREDICTED: bifunctional dihydrofolate reductase-thymidylate synthase-like isoform X4 [Vitis vinifera] |
| 17 |
Hb_013691_030 |
0.2660825748 |
- |
- |
hypothetical protein CISIN_1g020131mg [Citrus sinensis] |
| 18 |
Hb_005489_010 |
0.2664000997 |
- |
- |
hypothetical protein POPTR_0011s11420g [Populus trichocarpa] |
| 19 |
Hb_095192_010 |
0.2666993987 |
- |
- |
hypothetical protein VITISV_043980 [Vitis vinifera] |
| 20 |
Hb_182964_010 |
0.2683042493 |
- |
- |
PREDICTED: uncharacterized protein LOC105642044 isoform X2 [Jatropha curcas] |