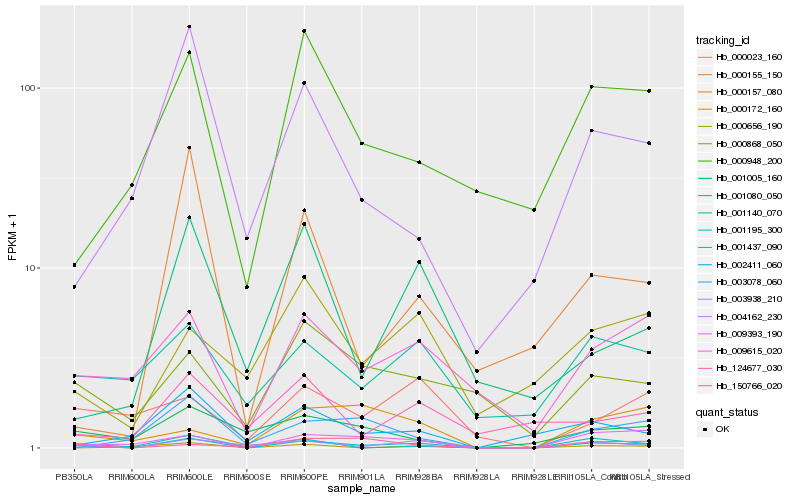
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001437_090 |
0.0 |
- |
- |
PREDICTED: phosphatidylinositol N-acetylglucosaminyltransferase gpi3 subunit [Jatropha curcas] |
| 2 |
Hb_003938_210 |
0.2249963061 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 3 |
Hb_009393_190 |
0.2486357371 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase AIP2-like [Jatropha curcas] |
| 4 |
Hb_009615_020 |
0.2716924959 |
transcription factor |
TF Family: HMG |
PREDICTED: high mobility group B protein 7-like [Jatropha curcas] |
| 5 |
Hb_001140_070 |
0.2954526196 |
- |
- |
Chromosome transmission fidelity protein 8, putative isoform 2 [Theobroma cacao] |
| 6 |
Hb_000948_200 |
0.2973036584 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 7 |
Hb_000868_050 |
0.2981216962 |
- |
- |
RAD51 homolog [Populus nigra] |
| 8 |
Hb_000656_190 |
0.3010549112 |
- |
- |
PREDICTED: probable nucleoside diphosphate kinase 5 [Jatropha curcas] |
| 9 |
Hb_001195_300 |
0.3060571708 |
- |
- |
PREDICTED: DUF21 domain-containing protein At1g47330 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002411_060 |
0.3064210858 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 5 [Jatropha curcas] |
| 11 |
Hb_000155_150 |
0.308739704 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 12 |
Hb_001080_050 |
0.3128861059 |
- |
- |
PREDICTED: uncharacterized protein LOC105635235 [Jatropha curcas] |
| 13 |
Hb_150766_020 |
0.3131032954 |
- |
- |
hypothetical protein VITISV_030841 [Vitis vinifera] |
| 14 |
Hb_000172_160 |
0.3145295375 |
- |
- |
hypothetical protein POPTR_0007s05770g [Populus trichocarpa] |
| 15 |
Hb_004162_230 |
0.3153202226 |
- |
- |
PREDICTED: histone H2B [Jatropha curcas] |
| 16 |
Hb_003078_060 |
0.3165032943 |
- |
- |
60S ribosomal protein L32, putative [Ricinus communis] |
| 17 |
Hb_000157_080 |
0.3173860108 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized N-acetyltransferase ycf52 [Jatropha curcas] |
| 18 |
Hb_001005_160 |
0.3174407434 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000023_160 |
0.318645817 |
- |
- |
PREDICTED: uncharacterized protein LOC105636423 isoform X1 [Jatropha curcas] |
| 20 |
Hb_124677_030 |
0.3232460643 |
- |
- |
PREDICTED: uncharacterized protein LOC105650473 [Jatropha curcas] |