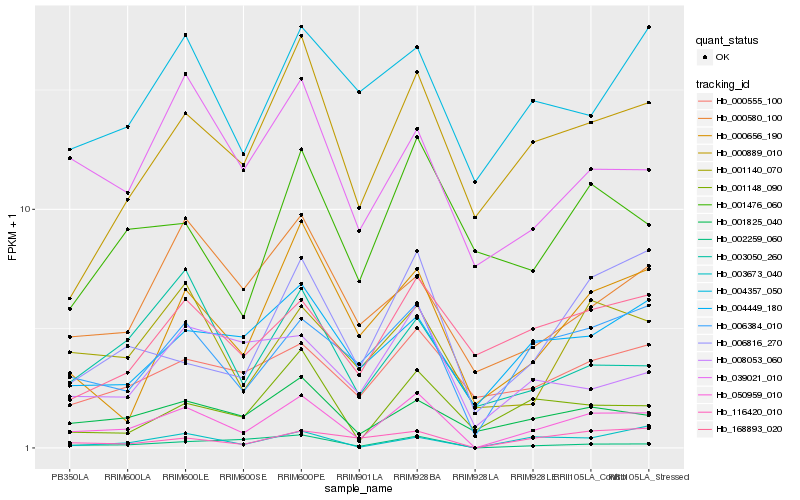
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_050959_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_006384_010 |
0.1543137007 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 3 |
Hb_001140_070 |
0.1796463105 |
- |
- |
Chromosome transmission fidelity protein 8, putative isoform 2 [Theobroma cacao] |
| 4 |
Hb_001825_040 |
0.1831005591 |
- |
- |
myosin XI, putative [Ricinus communis] |
| 5 |
Hb_116420_010 |
0.1840878984 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 6 |
Hb_002259_060 |
0.1879303014 |
- |
- |
PREDICTED: kinesin-like calmodulin-binding protein isoform X4 [Jatropha curcas] |
| 7 |
Hb_000889_010 |
0.1897933114 |
transcription factor |
TF Family: MYB-related |
hypothetical protein PRUPE_ppa014728mg [Prunus persica] |
| 8 |
Hb_004449_180 |
0.195676763 |
- |
- |
PREDICTED: SAC3 family protein 1 [Jatropha curcas] |
| 9 |
Hb_000656_190 |
0.1967979404 |
- |
- |
PREDICTED: probable nucleoside diphosphate kinase 5 [Jatropha curcas] |
| 10 |
Hb_006816_270 |
0.1972364094 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000555_100 |
0.1979023011 |
- |
- |
hypothetical protein JCGZ_14689 [Jatropha curcas] |
| 12 |
Hb_039021_010 |
0.2017503794 |
transcription factor |
TF Family: B3 |
hypothetical protein RCOM_0465210 [Ricinus communis] |
| 13 |
Hb_008053_060 |
0.202976418 |
- |
- |
PREDICTED: protein PAIR1 isoform X2 [Jatropha curcas] |
| 14 |
Hb_003673_040 |
0.206949018 |
- |
- |
dynamin family protein [Populus trichocarpa] |
| 15 |
Hb_001148_090 |
0.2077761335 |
- |
- |
hypothetical protein JCGZ_14711 [Jatropha curcas] |
| 16 |
Hb_000580_100 |
0.2078349475 |
- |
- |
PREDICTED: uncharacterized protein LOC105643133 isoform X1 [Jatropha curcas] |
| 17 |
Hb_004357_050 |
0.2106679429 |
- |
- |
PREDICTED: histone deacetylase HDT1-like [Jatropha curcas] |
| 18 |
Hb_001476_060 |
0.2111016286 |
- |
- |
PREDICTED: uncharacterized protein At5g01610 [Jatropha curcas] |
| 19 |
Hb_168893_020 |
0.2113415912 |
- |
- |
PREDICTED: probable protein S-acyltransferase 15 [Jatropha curcas] |
| 20 |
Hb_003050_260 |
0.2115377177 |
- |
- |
DNA replication licensing factor MCM3, putative [Ricinus communis] |