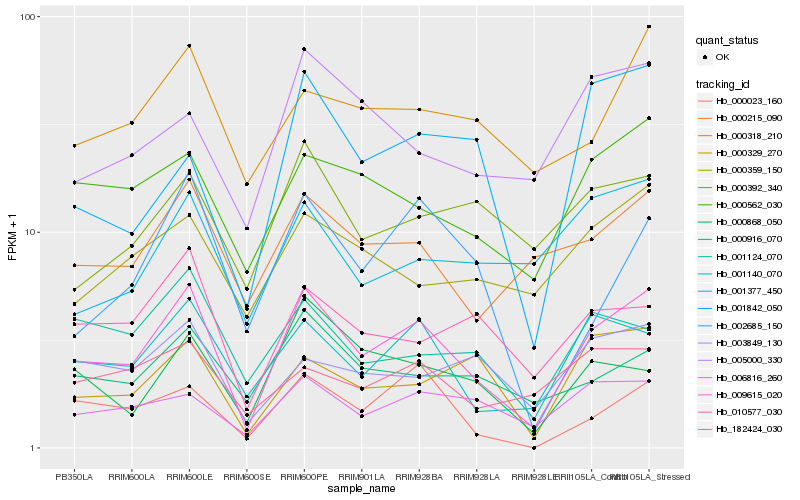
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009615_020 |
0.0 |
transcription factor |
TF Family: HMG |
PREDICTED: high mobility group B protein 7-like [Jatropha curcas] |
| 2 |
Hb_001140_070 |
0.1532832185 |
- |
- |
Chromosome transmission fidelity protein 8, putative isoform 2 [Theobroma cacao] |
| 3 |
Hb_006816_260 |
0.1660892149 |
- |
- |
hypothetical protein JCGZ_23244 [Jatropha curcas] |
| 4 |
Hb_000562_030 |
0.1690162167 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000023_160 |
0.175297309 |
- |
- |
PREDICTED: uncharacterized protein LOC105636423 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000329_270 |
0.1761597046 |
- |
- |
PREDICTED: spindle and kinetochore-associated protein 1 homolog [Jatropha curcas] |
| 7 |
Hb_000916_070 |
0.1779312442 |
- |
- |
PREDICTED: sister chromatid cohesion protein DCC1-like [Jatropha curcas] |
| 8 |
Hb_000215_090 |
0.1784068935 |
- |
- |
cytochrome b5 domain-containing family protein [Populus trichocarpa] |
| 9 |
Hb_010577_030 |
0.1794493457 |
- |
- |
PREDICTED: putative cyclin-B3-1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_003849_130 |
0.1841048652 |
- |
- |
PREDICTED: probable protein phosphatase 2C 14 [Jatropha curcas] |
| 11 |
Hb_002685_150 |
0.1854396095 |
- |
- |
PREDICTED: 2-aminoethanethiol dioxygenase-like [Jatropha curcas] |
| 12 |
Hb_000868_050 |
0.1860944621 |
- |
- |
RAD51 homolog [Populus nigra] |
| 13 |
Hb_000318_210 |
0.1863153776 |
- |
- |
PREDICTED: 40S ribosomal protein S12-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_001124_070 |
0.1867049628 |
- |
- |
Serine/threonine-protein kinase ULK4 [Gossypium arboreum] |
| 15 |
Hb_001842_050 |
0.1899881919 |
- |
- |
PREDICTED: protein BUD31 homolog 1-like [Jatropha curcas] |
| 16 |
Hb_182424_030 |
0.1905888946 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 8 [Vitis vinifera] |
| 17 |
Hb_000359_150 |
0.1920111587 |
- |
- |
PREDICTED: protein Mpv17-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_005000_330 |
0.1944287629 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001377_450 |
0.1959546701 |
- |
- |
PREDICTED: pyridoxine/pyridoxamine 5'-phosphate oxidase 2 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000392_340 |
0.1963901082 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 29 [Jatropha curcas] |