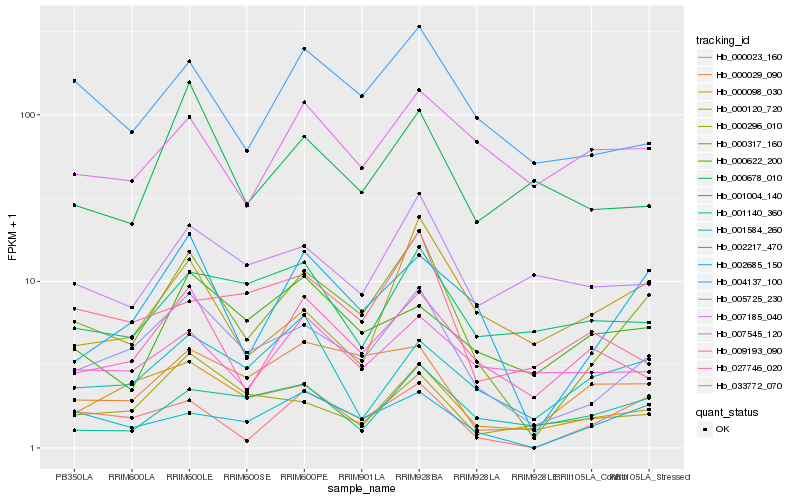
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000317_160 |
0.0 |
- |
- |
2-nonaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase [Theobroma cacao] |
| 2 |
Hb_005725_230 |
0.1160275042 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor ANT [Jatropha curcas] |
| 3 |
Hb_002685_150 |
0.1613793875 |
- |
- |
PREDICTED: 2-aminoethanethiol dioxygenase-like [Jatropha curcas] |
| 4 |
Hb_000023_160 |
0.1615559982 |
- |
- |
PREDICTED: uncharacterized protein LOC105636423 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002217_470 |
0.1685293806 |
- |
- |
PREDICTED: uncharacterized protein LOC105643559 [Jatropha curcas] |
| 6 |
Hb_004137_100 |
0.1839190833 |
- |
- |
PREDICTED: alcohol dehydrogenase 1 [Jatropha curcas] |
| 7 |
Hb_027746_020 |
0.1933415696 |
- |
- |
PREDICTED: replication protein A 70 kDa DNA-binding subunit B [Jatropha curcas] |
| 8 |
Hb_000029_090 |
0.1963310765 |
- |
- |
Nicalin precursor, putative [Ricinus communis] |
| 9 |
Hb_000098_030 |
0.2117780056 |
- |
- |
MLO-like protein 4 [Gossypium arboreum] |
| 10 |
Hb_007185_040 |
0.2175340098 |
- |
- |
PREDICTED: pyrroline-5-carboxylate reductase [Jatropha curcas] |
| 11 |
Hb_001140_360 |
0.2181026642 |
- |
- |
- |
| 12 |
Hb_000120_720 |
0.2223104281 |
transcription factor |
TF Family: HB |
homeobox-leucine zipper family protein [Populus trichocarpa] |
| 13 |
Hb_000678_010 |
0.2224060131 |
- |
- |
PREDICTED: ketol-acid reductoisomerase, chloroplastic-like [Jatropha curcas] |
| 14 |
Hb_001584_260 |
0.2241219346 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 15 |
Hb_007545_120 |
0.2248112085 |
- |
- |
NADH:cytochrome b5 reductase [Vernicia fordii] |
| 16 |
Hb_000296_010 |
0.2250738402 |
- |
- |
PREDICTED: TSL-kinase interacting protein 1 [Jatropha curcas] |
| 17 |
Hb_009193_090 |
0.2259333723 |
- |
- |
PREDICTED: protein root UVB sensitive 3 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000622_200 |
0.2273771715 |
- |
- |
aldose 1-epimerase, putative [Ricinus communis] |
| 19 |
Hb_001004_140 |
0.2274956472 |
- |
- |
hypothetical protein JCGZ_26893 [Jatropha curcas] |
| 20 |
Hb_033772_070 |
0.2291397942 |
- |
- |
PREDICTED: uncharacterized protein LOC105633681 [Jatropha curcas] |