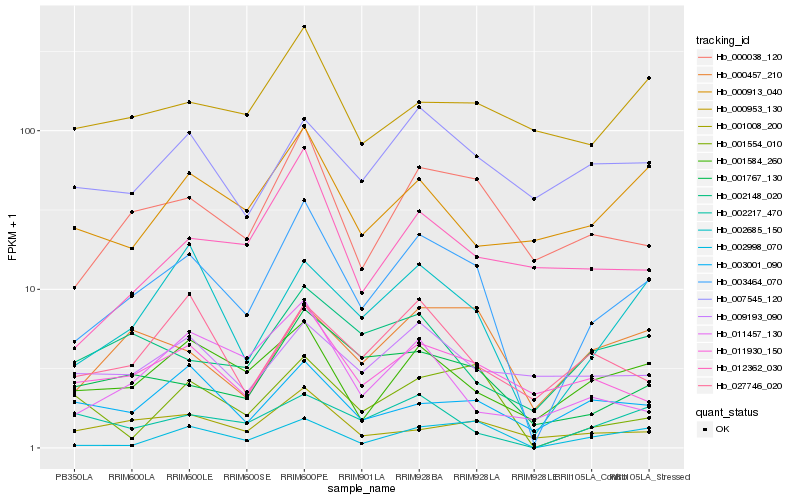
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003464_070 |
0.0 |
transcription factor |
TF Family: Trihelix |
PREDICTED: stress response protein NST1 [Jatropha curcas] |
| 2 |
Hb_000038_120 |
0.1635937678 |
- |
- |
PREDICTED: endoglucanase 9-like [Jatropha curcas] |
| 3 |
Hb_002685_150 |
0.1687643177 |
- |
- |
PREDICTED: 2-aminoethanethiol dioxygenase-like [Jatropha curcas] |
| 4 |
Hb_001767_130 |
0.1808758291 |
- |
- |
PREDICTED: beta-hexosaminidase 1 [Jatropha curcas] |
| 5 |
Hb_001584_260 |
0.1891072514 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 6 |
Hb_011930_150 |
0.1898734198 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002998_070 |
0.1961603994 |
- |
- |
hypothetical protein CARUB_v10019139mg, partial [Capsella rubella] |
| 8 |
Hb_000457_210 |
0.1991692359 |
- |
- |
PREDICTED: kelch repeat-containing protein At3g27220-like [Jatropha curcas] |
| 9 |
Hb_001008_200 |
0.2083873331 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC103961552 [Pyrus x bretschneideri] |
| 10 |
Hb_002217_470 |
0.2100524663 |
- |
- |
PREDICTED: uncharacterized protein LOC105643559 [Jatropha curcas] |
| 11 |
Hb_001554_010 |
0.2135750818 |
- |
- |
PREDICTED: telomere repeat-binding factor 4-like [Jatropha curcas] |
| 12 |
Hb_007545_120 |
0.2148395259 |
- |
- |
NADH:cytochrome b5 reductase [Vernicia fordii] |
| 13 |
Hb_027746_020 |
0.2152503856 |
- |
- |
PREDICTED: replication protein A 70 kDa DNA-binding subunit B [Jatropha curcas] |
| 14 |
Hb_002148_020 |
0.2162015001 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 15 |
Hb_012362_030 |
0.2178667223 |
- |
- |
PREDICTED: GPI-anchored protein LORELEI-like [Populus euphratica] |
| 16 |
Hb_000913_040 |
0.2207902169 |
- |
- |
Methyltransferase-related protein, putative [Theobroma cacao] |
| 17 |
Hb_000953_130 |
0.2217139974 |
- |
- |
PREDICTED: fructose-bisphosphate aldolase cytoplasmic isozyme [Jatropha curcas] |
| 18 |
Hb_009193_090 |
0.2223813599 |
- |
- |
PREDICTED: protein root UVB sensitive 3 isoform X1 [Jatropha curcas] |
| 19 |
Hb_003001_090 |
0.2229851583 |
- |
- |
PREDICTED: uncharacterized protein LOC105644365 [Jatropha curcas] |
| 20 |
Hb_011457_130 |
0.2232792938 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek2 [Jatropha curcas] |