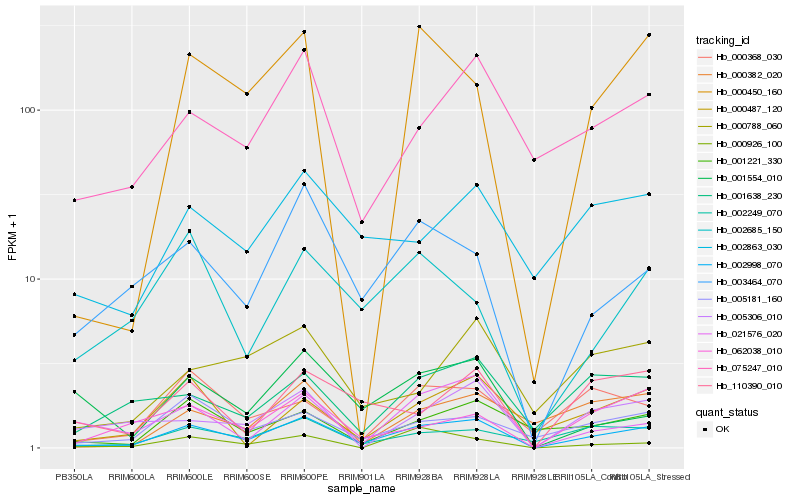
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002998_070 |
0.0 |
- |
- |
hypothetical protein CARUB_v10019139mg, partial [Capsella rubella] |
| 2 |
Hb_002249_070 |
0.1958343114 |
- |
- |
gcn4-complementing protein, putative [Ricinus communis] |
| 3 |
Hb_003464_070 |
0.1961603994 |
transcription factor |
TF Family: Trihelix |
PREDICTED: stress response protein NST1 [Jatropha curcas] |
| 4 |
Hb_005181_160 |
0.2045024993 |
- |
- |
PREDICTED: uncharacterized protein LOC105643546 [Jatropha curcas] |
| 5 |
Hb_075247_010 |
0.2047114501 |
- |
- |
PREDICTED: actin-depolymerizing factor 2-like [Nelumbo nucifera] |
| 6 |
Hb_000450_160 |
0.2108153859 |
- |
- |
hypothetical protein JCGZ_16746 [Jatropha curcas] |
| 7 |
Hb_001221_330 |
0.2123642174 |
- |
- |
PREDICTED: uncharacterized protein LOC105648791 [Jatropha curcas] |
| 8 |
Hb_021576_020 |
0.2140400425 |
- |
- |
PREDICTED: uncharacterized protein LOC104426132 [Eucalyptus grandis] |
| 9 |
Hb_110390_010 |
0.2145946948 |
- |
- |
hypothetical protein JCGZ_19741 [Jatropha curcas] |
| 10 |
Hb_001638_230 |
0.2160730705 |
- |
- |
RNA-binding region-containing protein, putative [Ricinus communis] |
| 11 |
Hb_062038_010 |
0.221111843 |
transcription factor |
TF Family: B3 |
PREDICTED: uncharacterized protein LOC104882266 isoform X2 [Vitis vinifera] |
| 12 |
Hb_002863_030 |
0.2229108195 |
- |
- |
unknown [Populus trichocarpa x Populus deltoides] |
| 13 |
Hb_005306_010 |
0.225120336 |
- |
- |
- |
| 14 |
Hb_000487_120 |
0.2253009441 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000368_030 |
0.2256640404 |
- |
- |
PREDICTED: cytokinin dehydrogenase 5 [Jatropha curcas] |
| 16 |
Hb_001554_010 |
0.2261865806 |
- |
- |
PREDICTED: telomere repeat-binding factor 4-like [Jatropha curcas] |
| 17 |
Hb_002685_150 |
0.2263593566 |
- |
- |
PREDICTED: 2-aminoethanethiol dioxygenase-like [Jatropha curcas] |
| 18 |
Hb_000788_060 |
0.2295806589 |
- |
- |
PREDICTED: uncharacterized protein LOC105629705 [Jatropha curcas] |
| 19 |
Hb_000926_100 |
0.2317944329 |
- |
- |
hypothetical protein B456_007G076800 [Gossypium raimondii] |
| 20 |
Hb_000382_020 |
0.2323413915 |
- |
- |
PREDICTED: uncharacterized protein LOC105633610 [Jatropha curcas] |