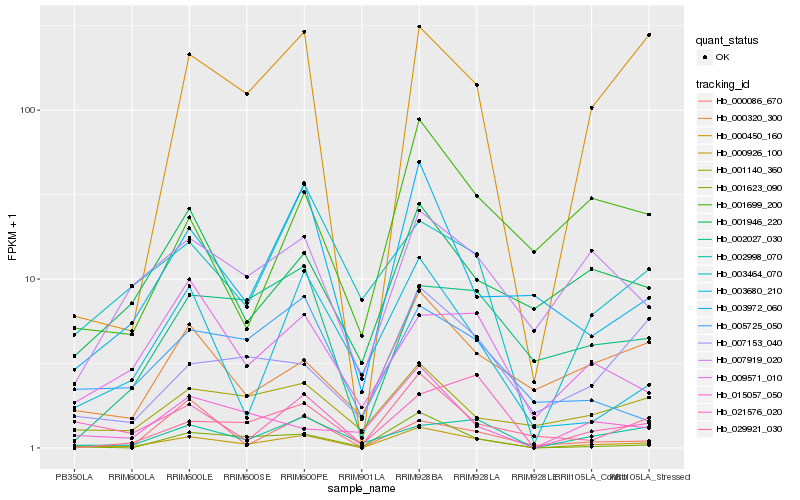
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000926_100 |
0.0 |
- |
- |
hypothetical protein B456_007G076800 [Gossypium raimondii] |
| 2 |
Hb_000450_160 |
0.213748622 |
- |
- |
hypothetical protein JCGZ_16746 [Jatropha curcas] |
| 3 |
Hb_001623_090 |
0.2169046845 |
- |
- |
PREDICTED: probable serine incorporator [Jatropha curcas] |
| 4 |
Hb_003972_060 |
0.2282911793 |
- |
- |
PREDICTED: UDP-arabinopyranose mutase 3 [Populus euphratica] |
| 5 |
Hb_029921_030 |
0.2303491225 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002998_070 |
0.2317944329 |
- |
- |
hypothetical protein CARUB_v10019139mg, partial [Capsella rubella] |
| 7 |
Hb_003680_210 |
0.2393435458 |
transcription factor |
TF Family: Trihelix |
PREDICTED: stress response protein NST1 [Jatropha curcas] |
| 8 |
Hb_000320_300 |
0.2424957752 |
- |
- |
prephenate dehydratase, putative [Ricinus communis] |
| 9 |
Hb_021576_020 |
0.247383908 |
- |
- |
PREDICTED: uncharacterized protein LOC104426132 [Eucalyptus grandis] |
| 10 |
Hb_007919_020 |
0.250438457 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 11 |
Hb_003464_070 |
0.2531005546 |
transcription factor |
TF Family: Trihelix |
PREDICTED: stress response protein NST1 [Jatropha curcas] |
| 12 |
Hb_002027_030 |
0.2531922823 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001140_360 |
0.2553161974 |
- |
- |
- |
| 14 |
Hb_001946_220 |
0.256477512 |
- |
- |
PREDICTED: signal peptidase complex catalytic subunit SEC11A-like [Jatropha curcas] |
| 15 |
Hb_009571_010 |
0.2565436732 |
- |
- |
early-responsive to dehydration family protein [Populus trichocarpa] |
| 16 |
Hb_005725_050 |
0.2583324451 |
- |
- |
PREDICTED: U-box domain-containing protein 30-like [Jatropha curcas] |
| 17 |
Hb_001699_200 |
0.2587613746 |
- |
- |
PREDICTED: chaperone protein dnaJ 11, chloroplastic [Jatropha curcas] |
| 18 |
Hb_007153_040 |
0.265126899 |
- |
- |
PREDICTED: uncharacterized protein LOC105633515 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000086_670 |
0.2655808847 |
- |
- |
PREDICTED: probable DNA helicase MCM9 [Jatropha curcas] |
| 20 |
Hb_015057_050 |
0.2705949626 |
- |
- |
D-alanine aminotransferase, putative [Ricinus communis] |