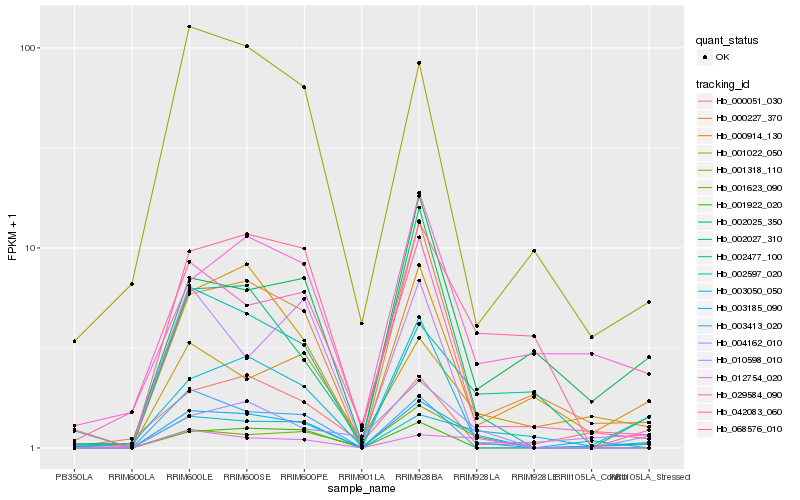
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003185_090 |
0.0 |
- |
- |
Boron transporter, putative [Ricinus communis] |
| 2 |
Hb_001623_090 |
0.1844478064 |
- |
- |
PREDICTED: probable serine incorporator [Jatropha curcas] |
| 3 |
Hb_012754_020 |
0.2020277393 |
- |
- |
PREDICTED: uncharacterized protein LOC104229837 [Nicotiana sylvestris] |
| 4 |
Hb_000227_370 |
0.202866219 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 5 |
Hb_001022_050 |
0.2079504582 |
- |
- |
hypothetical protein POPTR_0016s07780g [Populus trichocarpa] |
| 6 |
Hb_000914_130 |
0.2094800326 |
- |
- |
copper-transporting atpase p-type, putative [Ricinus communis] |
| 7 |
Hb_004162_010 |
0.2190339605 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_068576_010 |
0.2191914998 |
- |
- |
hypothetical protein JCGZ_21627 [Jatropha curcas] |
| 9 |
Hb_002477_100 |
0.2200282705 |
- |
- |
PREDICTED: protein Brevis radix-like 2 [Jatropha curcas] |
| 10 |
Hb_002597_020 |
0.2206696054 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 11 |
Hb_042083_060 |
0.2216888198 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g45780 [Jatropha curcas] |
| 12 |
Hb_003050_050 |
0.228671909 |
- |
- |
hypothetical protein JCGZ_04619 [Jatropha curcas] |
| 13 |
Hb_002027_310 |
0.2378970951 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001922_020 |
0.240406464 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_029584_090 |
0.240506819 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075 [Jatropha curcas] |
| 16 |
Hb_000051_030 |
0.2411618148 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase SINAT2-like [Jatropha curcas] |
| 17 |
Hb_003413_020 |
0.2439831177 |
- |
- |
- |
| 18 |
Hb_002025_350 |
0.2443205696 |
- |
- |
PREDICTED: glutamate decarboxylase 4 [Jatropha curcas] |
| 19 |
Hb_001318_110 |
0.2455054317 |
- |
- |
PREDICTED: uncharacterized protein LOC105642677 [Jatropha curcas] |
| 20 |
Hb_010598_010 |
0.2463544788 |
- |
- |
PREDICTED: ankyrin-1-like [Jatropha curcas] |