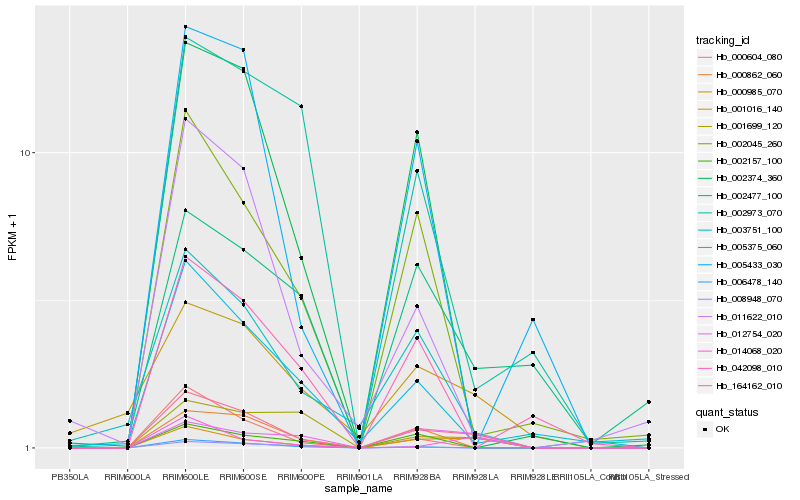
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_164162_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At1g08160-like [Jatropha curcas] |
| 2 |
Hb_000985_070 |
0.1570343892 |
- |
- |
PREDICTED: uncharacterized protein LOC105647984 [Jatropha curcas] |
| 3 |
Hb_012754_020 |
0.2195531145 |
- |
- |
PREDICTED: uncharacterized protein LOC104229837 [Nicotiana sylvestris] |
| 4 |
Hb_002045_260 |
0.2332056706 |
- |
- |
calmodulin, putative [Ricinus communis] |
| 5 |
Hb_006478_140 |
0.2352330201 |
- |
- |
PREDICTED: vicilin-like antimicrobial peptides 2-2 [Jatropha curcas] |
| 6 |
Hb_003751_100 |
0.2367592367 |
- |
- |
Amino acid permease 2 [Glycine soja] |
| 7 |
Hb_002374_360 |
0.2368932145 |
- |
- |
hypothetical protein POPTR_0002s08300g [Populus trichocarpa] |
| 8 |
Hb_000862_060 |
0.2401448664 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_011622_010 |
0.2407418494 |
- |
- |
PREDICTED: transcription factor DIVARICATA-like [Jatropha curcas] |
| 10 |
Hb_001016_140 |
0.2449656781 |
- |
- |
PREDICTED: uncharacterized protein LOC105647117 [Jatropha curcas] |
| 11 |
Hb_001699_120 |
0.247563291 |
- |
- |
Protein P21, putative [Ricinus communis] |
| 12 |
Hb_000604_080 |
0.2476048336 |
- |
- |
Fgenesh protein [Medicago truncatula] |
| 13 |
Hb_014068_020 |
0.2554595378 |
- |
- |
PREDICTED: uncharacterized protein LOC105628343 [Jatropha curcas] |
| 14 |
Hb_005375_060 |
0.2555613111 |
- |
- |
PREDICTED: probably inactive receptor-like protein kinase At2g46850 [Jatropha curcas] |
| 15 |
Hb_002157_100 |
0.2616404304 |
- |
- |
- |
| 16 |
Hb_005433_030 |
0.2649818633 |
transcription factor |
TF Family: MYB |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_042098_010 |
0.2711356175 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |
| 18 |
Hb_002477_100 |
0.2734478224 |
- |
- |
PREDICTED: protein Brevis radix-like 2 [Jatropha curcas] |
| 19 |
Hb_002973_070 |
0.2744144604 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 40 [Jatropha curcas] |
| 20 |
Hb_008948_070 |
0.2745853297 |
- |
- |
PREDICTED: phospholipase D alpha 1-like isoform X1 [Jatropha curcas] |