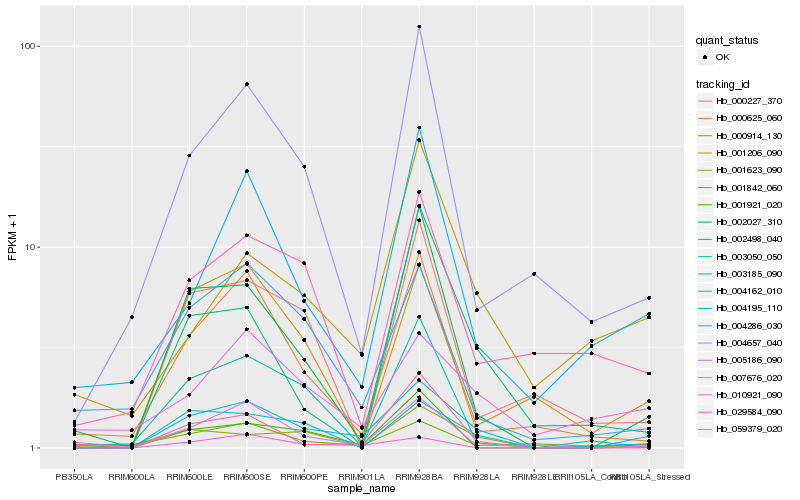
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004162_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001623_090 |
0.2121752603 |
- |
- |
PREDICTED: probable serine incorporator [Jatropha curcas] |
| 3 |
Hb_003185_090 |
0.2190339605 |
- |
- |
Boron transporter, putative [Ricinus communis] |
| 4 |
Hb_002027_310 |
0.2330408426 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_010921_090 |
0.2371399769 |
- |
- |
hypothetical protein JCGZ_04800 [Jatropha curcas] |
| 6 |
Hb_004657_040 |
0.2536773616 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor 15 family protein [Populus trichocarpa] |
| 7 |
Hb_004286_030 |
0.2598804191 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor ANT [Jatropha curcas] |
| 8 |
Hb_007676_020 |
0.2609728892 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase LIN-1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_005186_090 |
0.2623723679 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 10 |
Hb_004195_110 |
0.2642753233 |
- |
- |
hypothetical protein JCGZ_24010 [Jatropha curcas] |
| 11 |
Hb_000625_060 |
0.2681767447 |
- |
- |
Protein P21, putative [Ricinus communis] |
| 12 |
Hb_000914_130 |
0.2714798761 |
- |
- |
copper-transporting atpase p-type, putative [Ricinus communis] |
| 13 |
Hb_001921_020 |
0.2731044656 |
desease resistance |
Gene Name: LRR_8 |
PREDICTED: disease resistance protein RPM1-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_059379_020 |
0.273500363 |
- |
- |
PREDICTED: caffeic acid 3-O-methyltransferase 1-like [Populus euphratica] |
| 15 |
Hb_000227_370 |
0.2748094013 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 16 |
Hb_029584_090 |
0.2786646026 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075 [Jatropha curcas] |
| 17 |
Hb_001842_060 |
0.279129471 |
desease resistance |
Gene Name: ABC_trans_N |
hypothetical protein JCGZ_10908 [Jatropha curcas] |
| 18 |
Hb_001206_090 |
0.2792070797 |
- |
- |
pyruvate decarboxylase [Hevea brasiliensis] |
| 19 |
Hb_002498_040 |
0.2819829014 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 31 [Jatropha curcas] |
| 20 |
Hb_003050_050 |
0.2853368668 |
- |
- |
hypothetical protein JCGZ_04619 [Jatropha curcas] |