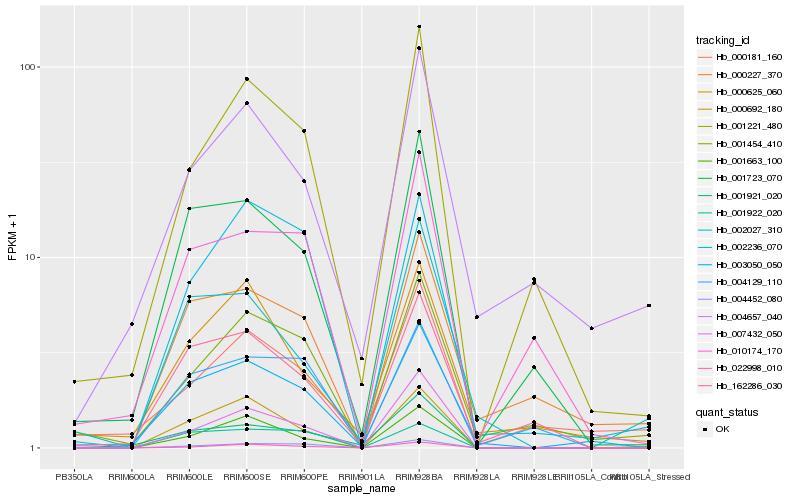
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003050_050 |
0.0 |
- |
- |
hypothetical protein JCGZ_04619 [Jatropha curcas] |
| 2 |
Hb_001723_070 |
0.1431016631 |
- |
- |
PREDICTED: polygalacturonase At1g48100 [Jatropha curcas] |
| 3 |
Hb_162286_030 |
0.1501689684 |
- |
- |
PREDICTED: uncharacterized protein LOC105650163 [Jatropha curcas] |
| 4 |
Hb_000692_180 |
0.1557782736 |
- |
- |
PREDICTED: uncharacterized protein At1g76070 [Jatropha curcas] |
| 5 |
Hb_000625_060 |
0.1561889126 |
- |
- |
Protein P21, putative [Ricinus communis] |
| 6 |
Hb_001221_480 |
0.1613559195 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 7 |
Hb_004452_080 |
0.1634745834 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 8 |
Hb_001454_410 |
0.1648571379 |
- |
- |
PREDICTED: uncharacterized protein LOC105643453 [Jatropha curcas] |
| 9 |
Hb_001663_100 |
0.1680980457 |
- |
- |
- |
| 10 |
Hb_000227_370 |
0.170020297 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 11 |
Hb_010174_170 |
0.172725435 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001921_020 |
0.1731291104 |
desease resistance |
Gene Name: LRR_8 |
PREDICTED: disease resistance protein RPM1-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_007432_050 |
0.1737356317 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB108 [Jatropha curcas] |
| 14 |
Hb_004657_040 |
0.1749006597 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor 15 family protein [Populus trichocarpa] |
| 15 |
Hb_002236_070 |
0.1756242013 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH36-like [Jatropha curcas] |
| 16 |
Hb_002027_310 |
0.1772150432 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001922_020 |
0.1792841249 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_022998_010 |
0.1804570621 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 19 |
Hb_004129_110 |
0.1814451457 |
- |
- |
Calcium-activated outward-rectifying potassium channel, putative [Ricinus communis] |
| 20 |
Hb_000181_160 |
0.1824147322 |
- |
- |
PREDICTED: uncharacterized protein LOC105637575 isoform X2 [Jatropha curcas] |