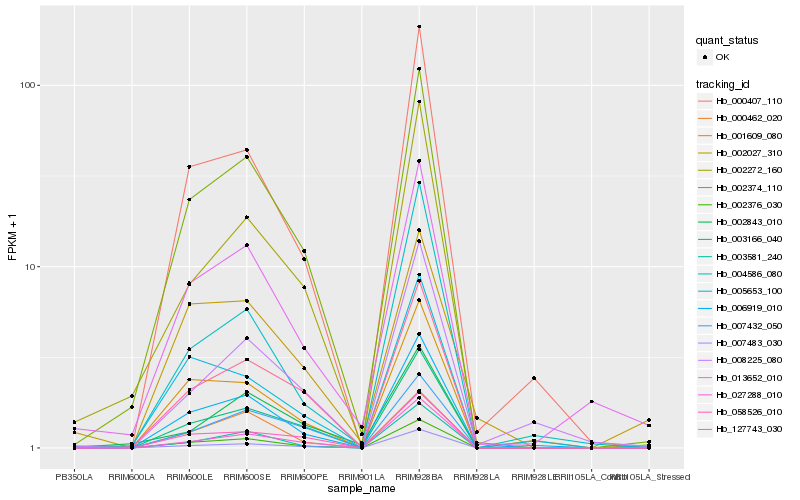
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_013652_010 |
0.0 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 2 |
Hb_002374_110 |
0.113858998 |
- |
- |
PREDICTED: uncharacterized protein LOC105638965 [Jatropha curcas] |
| 3 |
Hb_002376_030 |
0.1183360469 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_006919_010 |
0.1255241737 |
- |
- |
PREDICTED: putative wall-associated receptor kinase-like 16 [Vitis vinifera] |
| 5 |
Hb_000407_110 |
0.1470799152 |
- |
- |
ripening-induced ACC oxidase [Carica papaya] |
| 6 |
Hb_127743_030 |
0.1510825157 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 7 |
Hb_003166_040 |
0.1514330414 |
- |
- |
PREDICTED: probable indole-3-acetic acid-amido synthetase GH3.1 [Jatropha curcas] |
| 8 |
Hb_002272_160 |
0.1539041196 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 9 |
Hb_007483_030 |
0.1566690084 |
- |
- |
- |
| 10 |
Hb_001609_080 |
0.1584927312 |
- |
- |
hypothetical protein POPTR_0017s05660g [Populus trichocarpa] |
| 11 |
Hb_000462_020 |
0.1598518648 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 12 |
Hb_003581_240 |
0.1601308506 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 13 |
Hb_002027_310 |
0.1620492001 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_058526_010 |
0.1635332186 |
- |
- |
- |
| 15 |
Hb_027288_010 |
0.1636310912 |
- |
- |
mutt domain protein, putative [Ricinus communis] |
| 16 |
Hb_008225_080 |
0.1648834392 |
- |
- |
PREDICTED: cytochrome P450 71A26-like [Jatropha curcas] |
| 17 |
Hb_002843_010 |
0.1661647086 |
- |
- |
hypothetical protein EUGRSUZ_H04984, partial [Eucalyptus grandis] |
| 18 |
Hb_005653_100 |
0.1702671274 |
- |
- |
hypothetical protein POPTR_0004s19580g [Populus trichocarpa] |
| 19 |
Hb_004586_080 |
0.1731181319 |
- |
- |
hypothetical protein POPTR_0002s10520g [Populus trichocarpa] |
| 20 |
Hb_007432_050 |
0.1763594603 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB108 [Jatropha curcas] |