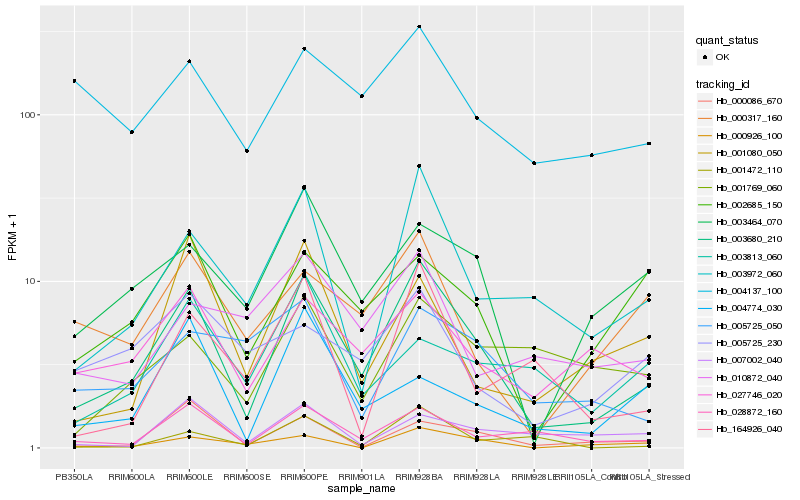
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003680_210 |
0.0 |
transcription factor |
TF Family: Trihelix |
PREDICTED: stress response protein NST1 [Jatropha curcas] |
| 2 |
Hb_003972_060 |
0.2007304919 |
- |
- |
PREDICTED: UDP-arabinopyranose mutase 3 [Populus euphratica] |
| 3 |
Hb_028872_160 |
0.2025753504 |
- |
- |
PREDICTED: ATP-dependent DNA helicase Q-like 5 [Jatropha curcas] |
| 4 |
Hb_027746_020 |
0.2243020572 |
- |
- |
PREDICTED: replication protein A 70 kDa DNA-binding subunit B [Jatropha curcas] |
| 5 |
Hb_003464_070 |
0.2270858018 |
transcription factor |
TF Family: Trihelix |
PREDICTED: stress response protein NST1 [Jatropha curcas] |
| 6 |
Hb_002685_150 |
0.2382604183 |
- |
- |
PREDICTED: 2-aminoethanethiol dioxygenase-like [Jatropha curcas] |
| 7 |
Hb_000926_100 |
0.2393435458 |
- |
- |
hypothetical protein B456_007G076800 [Gossypium raimondii] |
| 8 |
Hb_000317_160 |
0.2427170542 |
- |
- |
2-nonaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase [Theobroma cacao] |
| 9 |
Hb_000086_670 |
0.2428960948 |
- |
- |
PREDICTED: probable DNA helicase MCM9 [Jatropha curcas] |
| 10 |
Hb_001080_050 |
0.2432800622 |
- |
- |
PREDICTED: uncharacterized protein LOC105635235 [Jatropha curcas] |
| 11 |
Hb_164926_040 |
0.2453258272 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |
| 12 |
Hb_001472_110 |
0.2533778304 |
desease resistance |
Gene Name: TIR_2 |
nucleoside-triphosphatase, putative [Ricinus communis] |
| 13 |
Hb_004137_100 |
0.2546652962 |
- |
- |
PREDICTED: alcohol dehydrogenase 1 [Jatropha curcas] |
| 14 |
Hb_010872_040 |
0.2556008812 |
- |
- |
hypothetical protein F775_31993 [Aegilops tauschii] |
| 15 |
Hb_005725_050 |
0.2584404427 |
- |
- |
PREDICTED: U-box domain-containing protein 30-like [Jatropha curcas] |
| 16 |
Hb_005725_230 |
0.258686254 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor ANT [Jatropha curcas] |
| 17 |
Hb_004774_030 |
0.2595872735 |
- |
- |
PREDICTED: uncharacterized protein LOC105648177 [Jatropha curcas] |
| 18 |
Hb_001769_060 |
0.2603887427 |
- |
- |
PREDICTED: F-box protein At3g12350 isoform X1 [Jatropha curcas] |
| 19 |
Hb_007002_040 |
0.2619446302 |
transcription factor |
TF Family: PHD |
DNA replication regulator dpb11, putative [Ricinus communis] |
| 20 |
Hb_003813_060 |
0.2623648369 |
- |
- |
conserved hypothetical protein [Ricinus communis] |