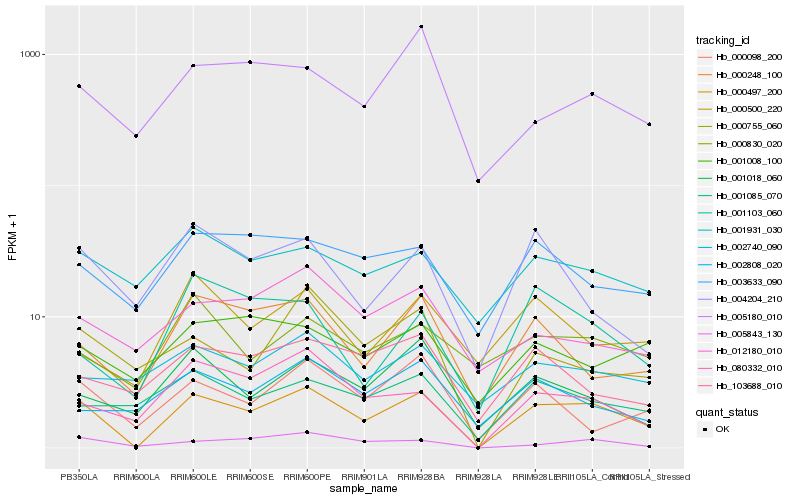
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000497_200 |
0.0 |
- |
- |
- |
| 2 |
Hb_005843_130 |
0.2103289843 |
- |
- |
hypothetical protein JCGZ_12389 [Jatropha curcas] |
| 3 |
Hb_080332_010 |
0.2131138293 |
- |
- |
hypothetical protein POPTR_0018s10590g [Populus trichocarpa] |
| 4 |
Hb_000248_100 |
0.2156373374 |
- |
- |
PREDICTED: NADPH-dependent thioredoxin reductase 3 [Populus euphratica] |
| 5 |
Hb_000098_200 |
0.2234015641 |
- |
- |
PREDICTED: serine hydroxymethyltransferase 7-like isoform X2 [Populus euphratica] |
| 6 |
Hb_001085_070 |
0.2267269714 |
- |
- |
PREDICTED: UBX domain-containing protein 4 [Vitis vinifera] |
| 7 |
Hb_000755_060 |
0.2287851672 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104424740 [Eucalyptus grandis] |
| 8 |
Hb_003633_090 |
0.229200091 |
- |
- |
arginine/serine-rich splicing factor, putative [Ricinus communis] |
| 9 |
Hb_001018_060 |
0.2303751316 |
- |
- |
hypothetical protein B456_001G028000 [Gossypium raimondii] |
| 10 |
Hb_001931_030 |
0.2344548661 |
- |
- |
PREDICTED: T-complex protein 1 subunit eta-like [Nelumbo nucifera] |
| 11 |
Hb_001103_060 |
0.2351122741 |
- |
- |
proton-dependent oligopeptide transport family protein [Populus trichocarpa] |
| 12 |
Hb_005180_010 |
0.2360517617 |
- |
- |
cyclophilin [Hevea brasiliensis] |
| 13 |
Hb_002740_090 |
0.2361518701 |
- |
- |
PREDICTED: putative membrane-bound O-acyltransferase C24H6.01c [Jatropha curcas] |
| 14 |
Hb_004204_210 |
0.2368184497 |
- |
- |
PREDICTED: heparanase-like protein 1 [Jatropha curcas] |
| 15 |
Hb_103688_010 |
0.2374169292 |
- |
- |
PREDICTED: sucrose-phosphatase 2-like [Jatropha curcas] |
| 16 |
Hb_002808_020 |
0.2385674937 |
- |
- |
PREDICTED: uncharacterized protein LOC105630318 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000830_020 |
0.2390244804 |
- |
- |
PREDICTED: 7-methylguanosine phosphate-specific 5'-nucleotidase A isoform X2 [Jatropha curcas] |
| 18 |
Hb_000500_220 |
0.2404901356 |
- |
- |
PREDICTED: intersectin-1 [Jatropha curcas] |
| 19 |
Hb_001008_100 |
0.2412895521 |
- |
- |
JHL25H03.10 [Jatropha curcas] |
| 20 |
Hb_012180_010 |
0.2414951014 |
- |
- |
PREDICTED: (S)-ureidoglycine aminohydrolase [Jatropha curcas] |